Pre Gene Modal
BGIBMGA005969
Annotation
PREDICTED:_cyclin-Y-like_protein_1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.742
Sequence
CDS
ATGGGTAATCAAAATAGTTGCTGCTGCTATCGGAGTCCATCTCCCATTCGCAAAGATATAGTAAAGTTGGAAGACTATCTTCCAGAAGGAGAAGTAAGCTCAAATAACATACAACACATCAGCGAGAGGGAGCCTGATGATGGCGACATCGATCCATCGCAAGATCCGATGGCTGGCACCATTTTCATGGAACGTTCTAAAGCTTCAATAGAGAATGGTATGACTAGGAAGCGGAGCCAACATCAAATAGCTGACAATAAATTAAAGAAAAGCAGTTCCTGTTCTACAATTTACTTAGATGATAGTACTGTGTCTCAACCAAATTTGAAAAACACTGTGAGGTGTGTAGCACTTGCTATTTACTATCACATAAAGAATAGAATGTCTGAGCGCAGGTTAGATATTTTTGATGAGAAACTTCATCCTTTGAGCAAAGAAGGTGTGAGTGATGACTATGATAAATACAATCCAGAGCATAAGCAGATTTATAAATTTGTAAGAACATTATTTAATGCAGCACAATTAACTGCAGAATGTGCTATTATTACTCTAGTCTATTTAGAGAGACTTCTAATTTGTGCTGAACTTGATATTGCACCTTCAAATTGGAAAAGAATTGTATTAGGGGCAATCTTATTAGCGAGTAAGGTATGGGACGATCAAGCAGTTTGGAATGTTGACTACTGTCAAATTCTAAAAGATATAACTGTTGAAGATATGAATGAGCTTGAACGACAATTTCTAGAAATGTTACAATTCAATATCAATGTTCCATCGAGTGTCTATGCTAAATACTATTTTGATCTACGTACACTTGCTGAAGCTAATGACCTAGCATTTCCTAGTGAGCCTCTTAGCAAGGAAAGGGCACAAAAGTTAGAAGCGATGTCACGAGTTATGGAAGACAAAATAACTGCTGAAATTGTAAAAAATGGTATTAAGAAATGGTCAAGTTTAGACAATGTTAATGCTGGTGCTGGAGTAAGACGAAGTGTAGCCATTTTGTCTTAA
Protein
MGNQNSCCCYRSPSPIRKDIVKLEDYLPEGEVSSNNIQHISEREPDDGDIDPSQDPMAGTIFMERSKASIENGMTRKRSQHQIADNKLKKSSSCSTIYLDDSTVSQPNLKNTVRCVALAIYYHIKNRMSERRLDIFDEKLHPLSKEGVSDDYDKYNPEHKQIYKFVRTLFNAAQLTAECAIITLVYLERLLICAELDIAPSNWKRIVLGAILLASKVWDDQAVWNVDYCQILKDITVEDMNELERQFLEMLQFNINVPSSVYAKYYFDLRTLAEANDLAFPSEPLSKERAQKLEAMSRVMEDKITAEIVKNGIKKWSSLDNVNAGAGVRRSVAILS
Summary
Similarity
Belongs to the cyclin family.
Uniprot
H9J8X7
A0A2A4JCW6
A0A2H1VYC9
S4PCS3
A0A194R2C9
A0A194PKT2
+ More
A0A212F618 A0A0L7KXU4 A0A3S2LT47 A0A1W4XK15 D6WMR1 A0A1Y1LQG5 N6U6G3 A0A023ERC2 Q16SP3 A0A195CHA3 Q7PWH0 A0A195E6S0 F4X6V9 A0A195F1B7 A0A158NFX2 E1ZYZ2 E9J657 E2BX36 A0A1Q3FU46 K7IQ09 A0A2M4AFZ5 A0A182NUP7 A0A2P8YTI9 A0A182F314 A0A2M4BSD9 W5JNA8 A0A182M946 A0A2M3Z2R4 A0A154P813 A0A182RZB1 A0A2A3E6K4 A0A087ZT32 A0A182PW10 A0A182WGB4 A0A084WQK3 A0A232F8K2 A0A182Q4H3 U5EYN6 A0A182LMK7 A0A182Y0H1 A0A195B2X0 A0A182J5T1 A0A1S4GZQ6 A0A182UMK5 A0A182UKN0 A0A182WXL5 A0A182I7H7 A0A0J7NGF3 A0A2J7Q0L5 A0A067RKG3 A0A336L890 A0A026WJ75 A0A0K8TL81 A0A069DT90 A0A0P4VU13 A0A0V0G7M7 R4G5I6 A0A2R7WPQ6 A0A1J1IKD6 A0A1L8DAW0 A0A0T6ASX2 A0A0L7RDX1 A0A1B6KLV5 A0A1B6CL35 A0A182T687 A0A2S2PER3 J9JZJ5 A0A1B0D187 A0A2H8TE60 A0A1I8Q1Q1 A0A0A1X4F2 W8CDC4 A0A0A9Z308 A0A0K8UKT3 A0A034VFR3 A0A0L0BLI9 A0A1B6GQR1 A0A1B6HN50 A0A3Q0J3N8 A0A1I8N7Y4 B4LQJ8 B4KIV7 A0A3B0JBW6 A0A0M4EQ77 B0XKN6 B4GPR4 Q29LM1 A0A1A9WXN4 B4JR29 B4P1J9 B4HX82 B4Q397 T1PJV2 A0A0M3QTR0
A0A212F618 A0A0L7KXU4 A0A3S2LT47 A0A1W4XK15 D6WMR1 A0A1Y1LQG5 N6U6G3 A0A023ERC2 Q16SP3 A0A195CHA3 Q7PWH0 A0A195E6S0 F4X6V9 A0A195F1B7 A0A158NFX2 E1ZYZ2 E9J657 E2BX36 A0A1Q3FU46 K7IQ09 A0A2M4AFZ5 A0A182NUP7 A0A2P8YTI9 A0A182F314 A0A2M4BSD9 W5JNA8 A0A182M946 A0A2M3Z2R4 A0A154P813 A0A182RZB1 A0A2A3E6K4 A0A087ZT32 A0A182PW10 A0A182WGB4 A0A084WQK3 A0A232F8K2 A0A182Q4H3 U5EYN6 A0A182LMK7 A0A182Y0H1 A0A195B2X0 A0A182J5T1 A0A1S4GZQ6 A0A182UMK5 A0A182UKN0 A0A182WXL5 A0A182I7H7 A0A0J7NGF3 A0A2J7Q0L5 A0A067RKG3 A0A336L890 A0A026WJ75 A0A0K8TL81 A0A069DT90 A0A0P4VU13 A0A0V0G7M7 R4G5I6 A0A2R7WPQ6 A0A1J1IKD6 A0A1L8DAW0 A0A0T6ASX2 A0A0L7RDX1 A0A1B6KLV5 A0A1B6CL35 A0A182T687 A0A2S2PER3 J9JZJ5 A0A1B0D187 A0A2H8TE60 A0A1I8Q1Q1 A0A0A1X4F2 W8CDC4 A0A0A9Z308 A0A0K8UKT3 A0A034VFR3 A0A0L0BLI9 A0A1B6GQR1 A0A1B6HN50 A0A3Q0J3N8 A0A1I8N7Y4 B4LQJ8 B4KIV7 A0A3B0JBW6 A0A0M4EQ77 B0XKN6 B4GPR4 Q29LM1 A0A1A9WXN4 B4JR29 B4P1J9 B4HX82 B4Q397 T1PJV2 A0A0M3QTR0
Pubmed
19121390
23622113
26354079
22118469
26227816
18362917
+ More
19820115 28004739 23537049 24945155 26483478 17510324 12364791 21719571 21347285 20798317 21282665 20075255 29403074 20920257 23761445 24438588 28648823 20966253 25244985 24845553 24508170 30249741 26369729 26334808 27129103 25830018 24495485 25401762 26823975 25348373 26108605 25315136 17994087 15632085 23185243 17550304 22936249
19820115 28004739 23537049 24945155 26483478 17510324 12364791 21719571 21347285 20798317 21282665 20075255 29403074 20920257 23761445 24438588 28648823 20966253 25244985 24845553 24508170 30249741 26369729 26334808 27129103 25830018 24495485 25401762 26823975 25348373 26108605 25315136 17994087 15632085 23185243 17550304 22936249
EMBL
BABH01004875
NWSH01001848
PCG69925.1
ODYU01004889
SOQ45224.1
GAIX01004011
+ More
JAA88549.1 KQ460855 KPJ11943.1 KQ459601 KPI93937.1 AGBW02010086 OWR49180.1 JTDY01004526 KOB68047.1 RSAL01000014 RVE53283.1 KQ971341 EFA03779.1 GEZM01053157 GEZM01053156 JAV74155.1 APGK01040724 KB740984 KB631904 ENN76246.1 ERL87075.1 JXUM01004990 JXUM01013405 GAPW01002402 KQ560376 KQ560185 JAC11196.1 KXJ82719.1 KXJ83942.1 CH477670 EAT37476.1 KQ977754 KYN00116.1 AAAB01008984 EAA14817.3 KQ979568 KYN20878.1 GL888818 EGI57891.1 KQ981864 KYN34258.1 ADTU01014825 GL435242 EFN73682.1 GL768221 EFZ11698.1 GL451202 EFN79755.1 GFDL01003958 JAV31087.1 GGFK01006375 MBW39696.1 PYGN01000369 PSN47554.1 GGFJ01006762 MBW55903.1 ADMH02000571 ETN65852.1 AXCM01019394 GGFM01002044 MBW22795.1 KQ434823 KZC07260.1 KZ288349 PBC27377.1 ATLV01025637 KE525396 KFB52497.1 NNAY01000723 OXU26789.1 AXCN02000736 GANO01000184 JAB59687.1 KQ976662 KYM78544.1 APCN01003406 LBMM01005312 KMQ91620.1 NEVH01019965 PNF22125.1 KK852415 KDR24361.1 UFQS01001999 UFQT01001999 SSX12913.1 SSX32355.1 KK107178 QOIP01000006 EZA56048.1 RLU21229.1 GDAI01002665 JAI14938.1 GBGD01002037 JAC86852.1 GDKW01000559 JAI56036.1 GECL01002674 JAP03450.1 ACPB03011334 ACPB03011335 GAHY01000530 JAA76980.1 KK855123 PTY21000.1 CVRI01000054 CRL00640.1 GFDF01010485 JAV03599.1 LJIG01022904 KRT78137.1 KQ414612 KOC69182.1 GEBQ01027535 JAT12442.1 GEDC01023162 JAS14136.1 GGMR01015255 MBY27874.1 ABLF02025311 AJVK01002452 AJVK01002453 AJVK01002454 GFXV01000574 MBW12379.1 GBXI01011824 GBXI01008739 GBXI01000420 JAD02468.1 JAD05553.1 JAD13872.1 GAMC01002191 GAMC01002190 JAC04365.1 GBHO01010099 GBHO01010098 GBHO01007419 GBHO01001207 GBHO01001205 GBHO01001202 GBHO01001200 GBHO01001198 GBRD01016720 GDHC01021735 JAG33505.1 JAG33506.1 JAG36185.1 JAG42397.1 JAG42399.1 JAG42402.1 JAG42404.1 JAG42406.1 JAG49107.1 JAP96893.1 GDHF01025030 GDHF01017700 JAI27284.1 JAI34614.1 GAKP01018000 GAKP01017996 JAC40952.1 JRES01001695 KNC20887.1 GECZ01004987 JAS64782.1 GECU01031602 JAS76104.1 CH940649 EDW64455.1 CH933807 EDW12463.1 OUUW01000004 SPP79857.1 CP012523 ALC39125.1 DS233926 EDS32355.1 CH479187 EDW39586.1 CH379060 EAL34024.1 KRT04404.1 CH916372 EDV99359.1 CM000157 EDW88106.1 CH480818 EDW52627.1 CM000361 CM002910 EDX04728.1 KMY89817.1 KA648375 AFP63004.1 ALC39343.1
JAA88549.1 KQ460855 KPJ11943.1 KQ459601 KPI93937.1 AGBW02010086 OWR49180.1 JTDY01004526 KOB68047.1 RSAL01000014 RVE53283.1 KQ971341 EFA03779.1 GEZM01053157 GEZM01053156 JAV74155.1 APGK01040724 KB740984 KB631904 ENN76246.1 ERL87075.1 JXUM01004990 JXUM01013405 GAPW01002402 KQ560376 KQ560185 JAC11196.1 KXJ82719.1 KXJ83942.1 CH477670 EAT37476.1 KQ977754 KYN00116.1 AAAB01008984 EAA14817.3 KQ979568 KYN20878.1 GL888818 EGI57891.1 KQ981864 KYN34258.1 ADTU01014825 GL435242 EFN73682.1 GL768221 EFZ11698.1 GL451202 EFN79755.1 GFDL01003958 JAV31087.1 GGFK01006375 MBW39696.1 PYGN01000369 PSN47554.1 GGFJ01006762 MBW55903.1 ADMH02000571 ETN65852.1 AXCM01019394 GGFM01002044 MBW22795.1 KQ434823 KZC07260.1 KZ288349 PBC27377.1 ATLV01025637 KE525396 KFB52497.1 NNAY01000723 OXU26789.1 AXCN02000736 GANO01000184 JAB59687.1 KQ976662 KYM78544.1 APCN01003406 LBMM01005312 KMQ91620.1 NEVH01019965 PNF22125.1 KK852415 KDR24361.1 UFQS01001999 UFQT01001999 SSX12913.1 SSX32355.1 KK107178 QOIP01000006 EZA56048.1 RLU21229.1 GDAI01002665 JAI14938.1 GBGD01002037 JAC86852.1 GDKW01000559 JAI56036.1 GECL01002674 JAP03450.1 ACPB03011334 ACPB03011335 GAHY01000530 JAA76980.1 KK855123 PTY21000.1 CVRI01000054 CRL00640.1 GFDF01010485 JAV03599.1 LJIG01022904 KRT78137.1 KQ414612 KOC69182.1 GEBQ01027535 JAT12442.1 GEDC01023162 JAS14136.1 GGMR01015255 MBY27874.1 ABLF02025311 AJVK01002452 AJVK01002453 AJVK01002454 GFXV01000574 MBW12379.1 GBXI01011824 GBXI01008739 GBXI01000420 JAD02468.1 JAD05553.1 JAD13872.1 GAMC01002191 GAMC01002190 JAC04365.1 GBHO01010099 GBHO01010098 GBHO01007419 GBHO01001207 GBHO01001205 GBHO01001202 GBHO01001200 GBHO01001198 GBRD01016720 GDHC01021735 JAG33505.1 JAG33506.1 JAG36185.1 JAG42397.1 JAG42399.1 JAG42402.1 JAG42404.1 JAG42406.1 JAG49107.1 JAP96893.1 GDHF01025030 GDHF01017700 JAI27284.1 JAI34614.1 GAKP01018000 GAKP01017996 JAC40952.1 JRES01001695 KNC20887.1 GECZ01004987 JAS64782.1 GECU01031602 JAS76104.1 CH940649 EDW64455.1 CH933807 EDW12463.1 OUUW01000004 SPP79857.1 CP012523 ALC39125.1 DS233926 EDS32355.1 CH479187 EDW39586.1 CH379060 EAL34024.1 KRT04404.1 CH916372 EDV99359.1 CM000157 EDW88106.1 CH480818 EDW52627.1 CM000361 CM002910 EDX04728.1 KMY89817.1 KA648375 AFP63004.1 ALC39343.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000283053 UP000192223 UP000007266 UP000019118 UP000030742 UP000069940 UP000249989 UP000008820 UP000078542 UP000007062 UP000078492 UP000007755 UP000078541 UP000005205 UP000000311 UP000008237 UP000002358 UP000075884 UP000245037 UP000069272 UP000000673 UP000075883 UP000076502 UP000075900 UP000242457 UP000005203 UP000075885 UP000075920 UP000030765 UP000215335 UP000075886 UP000075882 UP000076408 UP000078540 UP000075880 UP000075903 UP000075902 UP000076407 UP000075840 UP000036403 UP000235965 UP000027135 UP000053097 UP000279307 UP000015103 UP000183832 UP000053825 UP000075901 UP000007819 UP000092462 UP000095300 UP000037069 UP000079169 UP000095301 UP000008792 UP000009192 UP000268350 UP000092553 UP000002320 UP000008744 UP000001819 UP000091820 UP000001070 UP000002282 UP000001292 UP000000304
UP000283053 UP000192223 UP000007266 UP000019118 UP000030742 UP000069940 UP000249989 UP000008820 UP000078542 UP000007062 UP000078492 UP000007755 UP000078541 UP000005205 UP000000311 UP000008237 UP000002358 UP000075884 UP000245037 UP000069272 UP000000673 UP000075883 UP000076502 UP000075900 UP000242457 UP000005203 UP000075885 UP000075920 UP000030765 UP000215335 UP000075886 UP000075882 UP000076408 UP000078540 UP000075880 UP000075903 UP000075902 UP000076407 UP000075840 UP000036403 UP000235965 UP000027135 UP000053097 UP000279307 UP000015103 UP000183832 UP000053825 UP000075901 UP000007819 UP000092462 UP000095300 UP000037069 UP000079169 UP000095301 UP000008792 UP000009192 UP000268350 UP000092553 UP000002320 UP000008744 UP000001819 UP000091820 UP000001070 UP000002282 UP000001292 UP000000304
PRIDE
Pfam
PF00134 Cyclin_N
Interpro
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
H9J8X7
A0A2A4JCW6
A0A2H1VYC9
S4PCS3
A0A194R2C9
A0A194PKT2
+ More
A0A212F618 A0A0L7KXU4 A0A3S2LT47 A0A1W4XK15 D6WMR1 A0A1Y1LQG5 N6U6G3 A0A023ERC2 Q16SP3 A0A195CHA3 Q7PWH0 A0A195E6S0 F4X6V9 A0A195F1B7 A0A158NFX2 E1ZYZ2 E9J657 E2BX36 A0A1Q3FU46 K7IQ09 A0A2M4AFZ5 A0A182NUP7 A0A2P8YTI9 A0A182F314 A0A2M4BSD9 W5JNA8 A0A182M946 A0A2M3Z2R4 A0A154P813 A0A182RZB1 A0A2A3E6K4 A0A087ZT32 A0A182PW10 A0A182WGB4 A0A084WQK3 A0A232F8K2 A0A182Q4H3 U5EYN6 A0A182LMK7 A0A182Y0H1 A0A195B2X0 A0A182J5T1 A0A1S4GZQ6 A0A182UMK5 A0A182UKN0 A0A182WXL5 A0A182I7H7 A0A0J7NGF3 A0A2J7Q0L5 A0A067RKG3 A0A336L890 A0A026WJ75 A0A0K8TL81 A0A069DT90 A0A0P4VU13 A0A0V0G7M7 R4G5I6 A0A2R7WPQ6 A0A1J1IKD6 A0A1L8DAW0 A0A0T6ASX2 A0A0L7RDX1 A0A1B6KLV5 A0A1B6CL35 A0A182T687 A0A2S2PER3 J9JZJ5 A0A1B0D187 A0A2H8TE60 A0A1I8Q1Q1 A0A0A1X4F2 W8CDC4 A0A0A9Z308 A0A0K8UKT3 A0A034VFR3 A0A0L0BLI9 A0A1B6GQR1 A0A1B6HN50 A0A3Q0J3N8 A0A1I8N7Y4 B4LQJ8 B4KIV7 A0A3B0JBW6 A0A0M4EQ77 B0XKN6 B4GPR4 Q29LM1 A0A1A9WXN4 B4JR29 B4P1J9 B4HX82 B4Q397 T1PJV2 A0A0M3QTR0
A0A212F618 A0A0L7KXU4 A0A3S2LT47 A0A1W4XK15 D6WMR1 A0A1Y1LQG5 N6U6G3 A0A023ERC2 Q16SP3 A0A195CHA3 Q7PWH0 A0A195E6S0 F4X6V9 A0A195F1B7 A0A158NFX2 E1ZYZ2 E9J657 E2BX36 A0A1Q3FU46 K7IQ09 A0A2M4AFZ5 A0A182NUP7 A0A2P8YTI9 A0A182F314 A0A2M4BSD9 W5JNA8 A0A182M946 A0A2M3Z2R4 A0A154P813 A0A182RZB1 A0A2A3E6K4 A0A087ZT32 A0A182PW10 A0A182WGB4 A0A084WQK3 A0A232F8K2 A0A182Q4H3 U5EYN6 A0A182LMK7 A0A182Y0H1 A0A195B2X0 A0A182J5T1 A0A1S4GZQ6 A0A182UMK5 A0A182UKN0 A0A182WXL5 A0A182I7H7 A0A0J7NGF3 A0A2J7Q0L5 A0A067RKG3 A0A336L890 A0A026WJ75 A0A0K8TL81 A0A069DT90 A0A0P4VU13 A0A0V0G7M7 R4G5I6 A0A2R7WPQ6 A0A1J1IKD6 A0A1L8DAW0 A0A0T6ASX2 A0A0L7RDX1 A0A1B6KLV5 A0A1B6CL35 A0A182T687 A0A2S2PER3 J9JZJ5 A0A1B0D187 A0A2H8TE60 A0A1I8Q1Q1 A0A0A1X4F2 W8CDC4 A0A0A9Z308 A0A0K8UKT3 A0A034VFR3 A0A0L0BLI9 A0A1B6GQR1 A0A1B6HN50 A0A3Q0J3N8 A0A1I8N7Y4 B4LQJ8 B4KIV7 A0A3B0JBW6 A0A0M4EQ77 B0XKN6 B4GPR4 Q29LM1 A0A1A9WXN4 B4JR29 B4P1J9 B4HX82 B4Q397 T1PJV2 A0A0M3QTR0
Ontologies
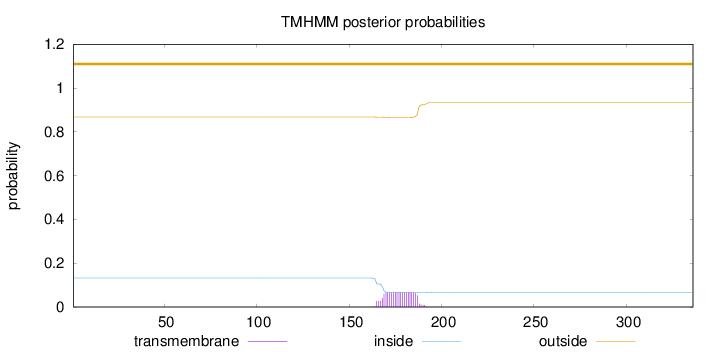
Topology
Length:
336
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.44487
Exp number, first 60 AAs:
0
Total prob of N-in:
0.13266
outside
1 - 336
Population Genetic Test Statistics
Pi
243.194958
Theta
207.69999
Tajima's D
0.415787
CLR
0.067325
CSRT
0.492275386230688
Interpretation
Uncertain
