Gene
KWMTBOMO01839
Pre Gene Modal
BGIBMGA005970
Annotation
PREDICTED:_uncharacterized_protein_LOC101741820_[Bombyx_mori]
Full name
XK-related protein
Location in the cell
PlasmaMembrane Reliability : 4.875
Sequence
CDS
ATGGTGACTTGTGAATTGGATAAGGATATTTCTGGCGAGGTTTTAGGTTGGAAGATCCCGGCATGGCAGGCGCTGTTGCTGAACAGAGTACTGCCTTCTTGCGGTGGGCTTATTGTTTATCTAATAGTCATATGTTTCGACTTAACATTAATTGTTGAGCATTTCCAAAACAATGACAACGGATTAGGAGCTATTTGCATAATTCTGATGATTTTGCCGTCGATAGTGTCAACGGTTTTCACCCTTGCCTCTCCCCCACCTGGCTTGGAAACTGAACTATCTGGATTCACGATAAATATTGAAAAAAATGATGTCAAATGGATTGTGATGCAGGTGGTCAACGCTATATTTTTTCCTATAGCAGCTATAGGGAGATATTGTTTCTTGATATTTTGGTGGGTGGAAGCTGTATGTGGTTCGAGGGCGGGTGACAAAGAAAGAACGCAAGAAGCGATACAGTTAGCTAGGGCTTCGTCTCCAATGGAACTCTATCTATTTCTCCAGTCATTCATACATTCAGCTCCACACGCAATTATTAATATCTTGTATCTGATGTCGAGATTTAATGACATAACGTACGGCAAAGTATCGGTTCAATCAATAAGTATAATCGCTGCTTCTCTTCGCATGGCCAGTACTGCTACAATTTACAGACGGTTCGAAAGAGAAAAGTTATGCGGTCGCAAATATCCTTGGAGTAAAAATGTTGAACAGGATCCTTTAATTCATTCAAACGCAAACGTAATGCAGAAACAAATACCTGAAGATCTTTCGGAAAATATTTATGAAGCCGTTCCCAAAAGTGAAACTACATATTCTGACTCCAGAAATCCAGTTAAAACAAACAGCGATCTAATACAGTTCTCGCCGAGGAACACTTACGAATCCAATTTTTATGATGACATTTCAACAGATTCACATCTTAATTTTGATTATATATCATCGGTGCCATCGTATGGATATTCTCAATCGTCGTCGGTTGATAGCGATGAAGAGTACGTCAAACCTGTTTCGATTATAGACCGGGTGGCACCGAGACGTCGGGGCACAGAATACAATATAGAAAGAGTAAATATAACTCCACCTCCAATAATACCCGCGCCAAGGCCGGGCTCCATCGCTGTTTGGGCTGAGAAGATTGTTGAAAATGCCGAATCTATACCAAGCTGGCTCTCCGCTCCACCACGGCGAAAATACTGCGAAGAAGTGATCAAAGAAGAGCCCGACATTCCCCGTAAGCTACCCATAACATACATGCGAGGTTTGGAGCCACAAGACACCACTGCCGCCTTGGTTAACTTTTTGGGTTGGTACTCGTTCTTTATAATGCTGTTATTCTTGATAGTTCCTCAGGCGAGTACCGTGAAGCGAGGATTCTACTTGTTCCTAGCGTTTATCTACTTATTCTGTTTGATGGAGTTCAAGATACGTTTCCGGCACGTGCGCGTGTGGCATGTGTTTTGGATTATTGTGTGTACTACAGAGACGGTTGTATTCACGGGAGTCTGGGCCTCTATAGACAATACGATGGAGTGTATTTTGTGTTATTGCAACCGCGGGAAACTATTGTTTACTTTAAGAATAAAAATAATAGTACGTAATACCAAGAACAATGTTTCATGA
Protein
MVTCELDKDISGEVLGWKIPAWQALLLNRVLPSCGGLIVYLIVICFDLTLIVEHFQNNDNGLGAICIILMILPSIVSTVFTLASPPPGLETELSGFTINIEKNDVKWIVMQVVNAIFFPIAAIGRYCFLIFWWVEAVCGSRAGDKERTQEAIQLARASSPMELYLFLQSFIHSAPHAIINILYLMSRFNDITYGKVSVQSISIIAASLRMASTATIYRRFEREKLCGRKYPWSKNVEQDPLIHSNANVMQKQIPEDLSENIYEAVPKSETTYSDSRNPVKTNSDLIQFSPRNTYESNFYDDISTDSHLNFDYISSVPSYGYSQSSSVDSDEEYVKPVSIIDRVAPRRRGTEYNIERVNITPPPIIPAPRPGSIAVWAEKIVENAESIPSWLSAPPRRKYCEEVIKEEPDIPRKLPITYMRGLEPQDTTAALVNFLGWYSFFIMLLFLIVPQASTVKRGFYLFLAFIYLFCLMEFKIRFRHVRVWHVFWIIVCTTETVVFTGVWASIDNTMECILCYCNRGKLLFTLRIKIIVRNTKNNVS
Summary
Similarity
Belongs to the XK family.
Uniprot
EMBL
Proteomes
Pfam
PF09815 XK-related
Interpro
IPR018629
XK-rel
ProteinModelPortal
Ontologies
GO
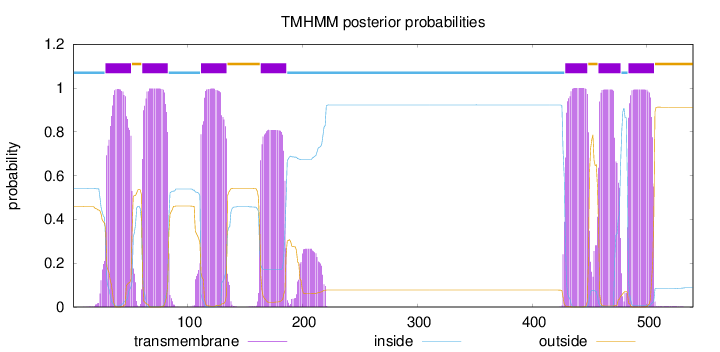
Topology
Subcellular location
Membrane
Length:
540
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
153.11285
Exp number, first 60 AAs:
22.04923
Total prob of N-in:
0.54110
POSSIBLE N-term signal
sequence
inside
1 - 28
TMhelix
29 - 51
outside
52 - 60
TMhelix
61 - 83
inside
84 - 111
TMhelix
112 - 134
outside
135 - 163
TMhelix
164 - 186
inside
187 - 428
TMhelix
429 - 448
outside
449 - 457
TMhelix
458 - 477
inside
478 - 483
TMhelix
484 - 506
outside
507 - 540
Population Genetic Test Statistics
Pi
287.156917
Theta
246.332744
Tajima's D
0.221499
CLR
0.016584
CSRT
0.430728463576821
Interpretation
Uncertain
