Gene
KWMTBOMO01838
Pre Gene Modal
BGIBMGA005972
Annotation
PREDICTED:_tyrosine-protein_kinase_transmembrane_receptor_Ror-like_isoform_X1_[Bombyx_mori]
Full name
Tyrosine-protein kinase receptor
Location in the cell
Nuclear Reliability : 3.653
Sequence
CDS
ATGCGTGCAAGGCATCGGGGCCATGAAGAAAATCTGACGAGGTTTGCCAGAGATGAATTGAATCTGGAAGAAAATTCGGAGCCACGACCAATTAAATACTTTCAAGGAGCTTGCGAGCCGATGTCCTGTGGAGAAGAAGCTGCGTGTACGGAATACAACAGTACCGTTCATTTCTGTTGGTGCACGACCACCGGAATGCCCATTACTGAGGACTTGCAATGCCCAAGAAGTCCAGTGACGGCAACTCCTAAACCGATCCATAATGTGATTCCGCCGCAGACTCGCACCACTAACACATTCGACGGAAGAAACGCGCCTTCACCGCCAACTGCTCAATTCAGTGGAGAAGTTTTGGCGGAAGCAACGACTGGAAATGAACCAAATGTTGTGGGAGCCGGTGTACACGTTACAACAGGAGTACTTCTGGCAGCTCTAGTTAGTGGAGCATTACTTCTTTTGTTCGTGGTATCTATTCTAGTTTGGTACTTTAGACGAAGGATATGTCGCGTCGATGGAAAAACTCAGCCGCGAGTGCGGCCAGGAGAGCCTCTTCTCGCTGAAAGGTATATAACGAACCCCCAGTACGGTGTGTACGGAGCCGGCGCGACCGCGACCGGCACGGGTCCGGCGAACGGACCGATTGTCGACGACACTACAGAACCCAGAGTACCTCTCTTAGATAGAAGCGCTTTGACATTCCTCGAAGAAGTCGGTGAAGGATGTTTTGGCAAGGTGCACAGAGGTTTGCTTCGAGTAAATGACTTGGAACAAGAAGTCGCAATTAAGGTTCTGAAAGAAAGCGCTGGACGCAGCGCCGAAGAGGAATTCATACGAGAGGTTTCAATAATGTCTGCATTTAGACATTCAAATATTCTTACTCTAGTCGGAGTGGTCTATAAAGATAATGTTATATGCAGCCCTTGGATGGTGTTTGAATATATGGAGCTGGGTGATTTGGCCGGTGTCCTGAGGGGGTCTCGCAATGGTGTTCGACCTGCACCCACGCTGAACGAAGTTGCTCTCTTACACGTAGCGCTACAAGTTGCGAGGGGCATGCAATACCTCGCTTCCCGCAGATTTGTCCACCGTGATTTAGCTGCCAGAAACTGTTTGGTCGGATCTCATCTTACAGTTAAGATAGCTGACTTTGGAATGTCCAGAGACGTTTACACTTGTGATTATTACAAAATGGGCGGTGAACGTCCAATGCCAGTACGGTGGATGTCTCCGGAGAGCATTCTGTACGCACGTTTTACTCACGAATCAGACGTGTGGGCGTATGGTGTCGTACTCTGGGAGATATACAGCCGAGGCAAACAACCATTTTATGGGCATAATAACGAAGATGCCACAAAGCTAATCGTACAGGGTATTTTGTTGGTGCCGCCAGAAGAATGTCCTCGGTTTGCTTGCGAGTTGATGCGCGAATGCTGGCAGTTCGATCCACAAAATAGGATCGATTTTGATGAGATTTGTAGAAAATTAGAAATAGCTGCGGCCGGTGGTACTACGACCGAGCAAGTGCGACTGCCGCGACCACCAAATCCTTCTCAAGATTCTGCTGGGTATCTGATCCCGGCGCGGCAAACTCCAGTTGATTATCTGACTTTATTGTTAGGTGATGAAAATCATGAGCCCGAATCTGAGTCTAGTGAAGAAGAGGAAGACGAAGAAGAGTATACTTGA
Protein
MRARHRGHEENLTRFARDELNLEENSEPRPIKYFQGACEPMSCGEEAACTEYNSTVHFCWCTTTGMPITEDLQCPRSPVTATPKPIHNVIPPQTRTTNTFDGRNAPSPPTAQFSGEVLAEATTGNEPNVVGAGVHVTTGVLLAALVSGALLLLFVVSILVWYFRRRICRVDGKTQPRVRPGEPLLAERYITNPQYGVYGAGATATGTGPANGPIVDDTTEPRVPLLDRSALTFLEEVGEGCFGKVHRGLLRVNDLEQEVAIKVLKESAGRSAEEEFIREVSIMSAFRHSNILTLVGVVYKDNVICSPWMVFEYMELGDLAGVLRGSRNGVRPAPTLNEVALLHVALQVARGMQYLASRRFVHRDLAARNCLVGSHLTVKIADFGMSRDVYTCDYYKMGGERPMPVRWMSPESILYARFTHESDVWAYGVVLWEIYSRGKQPFYGHNNEDATKLIVQGILLVPPEECPRFACELMRECWQFDPQNRIDFDEICRKLEIAAAGGTTTEQVRLPRPPNPSQDSAGYLIPARQTPVDYLTLLLGDENHEPESESSEEEEDEEEYT
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family. Insulin receptor subfamily.
Belongs to the protein kinase superfamily. Tyr protein kinase family. Insulin receptor subfamily.
Uniprot
H9J8Y0
A0A194R3X1
A0A194PKH6
A0A3S2NKJ8
A0A2A4KAF4
A0A212F626
+ More
A0A232EGX7 A0A1B6CKF4 A0A1B6KVD3 K7IPU6 A0A0L7R180 T1IAV2 A0A067RMT3 E2BLZ5 A0A0C9R5T8 A0A195BWX7 A0A195F1B4 A0A151IH99 A0A154NY25 A0A158P3J9 A0A2A3EGD4 A0A1Y1M813 A0A088ANR5 A0A1W4XB69 A0A151XE00 A0A0C9PQ15 F4WAT0 E9IEA7 A0A2P8YTG8 A0A224XQN1 A0A2H8TK98 A0A2R4G8U5 A0A0A9XGS5 A0A0K8ST81 J9JNV6 A0A2R7WVB6 A0A151IV50 A0A164UE11 A0A0N8CXK0 A0A0P5NAT6 A0A0N8BIW8 T1KK75 A0A0P6DY13 A0A0P6B4T2 A0A0P5T1Y6 A0A226F556 A0A0P4Y789
A0A232EGX7 A0A1B6CKF4 A0A1B6KVD3 K7IPU6 A0A0L7R180 T1IAV2 A0A067RMT3 E2BLZ5 A0A0C9R5T8 A0A195BWX7 A0A195F1B4 A0A151IH99 A0A154NY25 A0A158P3J9 A0A2A3EGD4 A0A1Y1M813 A0A088ANR5 A0A1W4XB69 A0A151XE00 A0A0C9PQ15 F4WAT0 E9IEA7 A0A2P8YTG8 A0A224XQN1 A0A2H8TK98 A0A2R4G8U5 A0A0A9XGS5 A0A0K8ST81 J9JNV6 A0A2R7WVB6 A0A151IV50 A0A164UE11 A0A0N8CXK0 A0A0P5NAT6 A0A0N8BIW8 T1KK75 A0A0P6DY13 A0A0P6B4T2 A0A0P5T1Y6 A0A226F556 A0A0P4Y789
EC Number
2.7.10.1
Pubmed
EMBL
BABH01004867
KQ460855
KPJ11940.1
KQ459601
KPI93941.1
RSAL01000014
+ More
RVE53286.1 NWSH01000024 PCG80632.1 AGBW02010086 OWR49183.1 NNAY01004651 OXU17605.1 GEDC01023445 JAS13853.1 GEBQ01024584 JAT15393.1 KQ414668 KOC64551.1 ACPB03006705 KK852415 KDR24363.1 GL449088 EFN83281.1 GBYB01003294 GBYB01003296 GBYB01003300 JAG73061.1 JAG73063.1 JAG73067.1 KQ976394 KYM93134.1 KQ981880 KYN33894.1 KQ977636 KYN01139.1 KQ434782 KZC04523.1 ADTU01008164 KZ288254 PBC30768.1 GEZM01041377 JAV80465.1 KQ982268 KYQ58478.1 GBYB01003293 GBYB01003295 JAG73060.1 JAG73062.1 GL888053 EGI68686.1 GL762576 EFZ21082.1 PYGN01000369 PSN47551.1 GFTR01006147 JAW10279.1 GFXV01002750 MBW14555.1 MF620046 AVT56268.1 GBHO01029468 GBHO01024450 GDHC01017328 JAG14136.1 JAG19154.1 JAQ01301.1 GBRD01009789 JAG56035.1 ABLF02023652 ABLF02023653 ABLF02023656 ABLF02023657 KK855629 PTY23341.1 KQ980924 KYN11407.1 LRGB01001581 KZS11277.1 GDIP01089047 JAM14668.1 GDIQ01155674 JAK96051.1 GDIQ01173315 JAK78410.1 CAEY01000173 CAEY01000174 CAEY01000175 GDIQ01071443 JAN23294.1 GDIP01020165 JAM83550.1 GDIP01132200 LRGB01001361 JAL71514.1 KZS12229.1 LNIX01000001 OXA64587.1 GDIP01232573 JAI90828.1
RVE53286.1 NWSH01000024 PCG80632.1 AGBW02010086 OWR49183.1 NNAY01004651 OXU17605.1 GEDC01023445 JAS13853.1 GEBQ01024584 JAT15393.1 KQ414668 KOC64551.1 ACPB03006705 KK852415 KDR24363.1 GL449088 EFN83281.1 GBYB01003294 GBYB01003296 GBYB01003300 JAG73061.1 JAG73063.1 JAG73067.1 KQ976394 KYM93134.1 KQ981880 KYN33894.1 KQ977636 KYN01139.1 KQ434782 KZC04523.1 ADTU01008164 KZ288254 PBC30768.1 GEZM01041377 JAV80465.1 KQ982268 KYQ58478.1 GBYB01003293 GBYB01003295 JAG73060.1 JAG73062.1 GL888053 EGI68686.1 GL762576 EFZ21082.1 PYGN01000369 PSN47551.1 GFTR01006147 JAW10279.1 GFXV01002750 MBW14555.1 MF620046 AVT56268.1 GBHO01029468 GBHO01024450 GDHC01017328 JAG14136.1 JAG19154.1 JAQ01301.1 GBRD01009789 JAG56035.1 ABLF02023652 ABLF02023653 ABLF02023656 ABLF02023657 KK855629 PTY23341.1 KQ980924 KYN11407.1 LRGB01001581 KZS11277.1 GDIP01089047 JAM14668.1 GDIQ01155674 JAK96051.1 GDIQ01173315 JAK78410.1 CAEY01000173 CAEY01000174 CAEY01000175 GDIQ01071443 JAN23294.1 GDIP01020165 JAM83550.1 GDIP01132200 LRGB01001361 JAL71514.1 KZS12229.1 LNIX01000001 OXA64587.1 GDIP01232573 JAI90828.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000218220
UP000007151
+ More
UP000215335 UP000002358 UP000053825 UP000015103 UP000027135 UP000008237 UP000078540 UP000078541 UP000078542 UP000076502 UP000005205 UP000242457 UP000005203 UP000192223 UP000075809 UP000007755 UP000245037 UP000007819 UP000078492 UP000076858 UP000015104 UP000198287
UP000215335 UP000002358 UP000053825 UP000015103 UP000027135 UP000008237 UP000078540 UP000078541 UP000078542 UP000076502 UP000005205 UP000242457 UP000005203 UP000192223 UP000075809 UP000007755 UP000245037 UP000007819 UP000078492 UP000076858 UP000015104 UP000198287
PRIDE
Pfam
PF07714 Pkinase_Tyr
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9J8Y0
A0A194R3X1
A0A194PKH6
A0A3S2NKJ8
A0A2A4KAF4
A0A212F626
+ More
A0A232EGX7 A0A1B6CKF4 A0A1B6KVD3 K7IPU6 A0A0L7R180 T1IAV2 A0A067RMT3 E2BLZ5 A0A0C9R5T8 A0A195BWX7 A0A195F1B4 A0A151IH99 A0A154NY25 A0A158P3J9 A0A2A3EGD4 A0A1Y1M813 A0A088ANR5 A0A1W4XB69 A0A151XE00 A0A0C9PQ15 F4WAT0 E9IEA7 A0A2P8YTG8 A0A224XQN1 A0A2H8TK98 A0A2R4G8U5 A0A0A9XGS5 A0A0K8ST81 J9JNV6 A0A2R7WVB6 A0A151IV50 A0A164UE11 A0A0N8CXK0 A0A0P5NAT6 A0A0N8BIW8 T1KK75 A0A0P6DY13 A0A0P6B4T2 A0A0P5T1Y6 A0A226F556 A0A0P4Y789
A0A232EGX7 A0A1B6CKF4 A0A1B6KVD3 K7IPU6 A0A0L7R180 T1IAV2 A0A067RMT3 E2BLZ5 A0A0C9R5T8 A0A195BWX7 A0A195F1B4 A0A151IH99 A0A154NY25 A0A158P3J9 A0A2A3EGD4 A0A1Y1M813 A0A088ANR5 A0A1W4XB69 A0A151XE00 A0A0C9PQ15 F4WAT0 E9IEA7 A0A2P8YTG8 A0A224XQN1 A0A2H8TK98 A0A2R4G8U5 A0A0A9XGS5 A0A0K8ST81 J9JNV6 A0A2R7WVB6 A0A151IV50 A0A164UE11 A0A0N8CXK0 A0A0P5NAT6 A0A0N8BIW8 T1KK75 A0A0P6DY13 A0A0P6B4T2 A0A0P5T1Y6 A0A226F556 A0A0P4Y789
Ontologies
GO
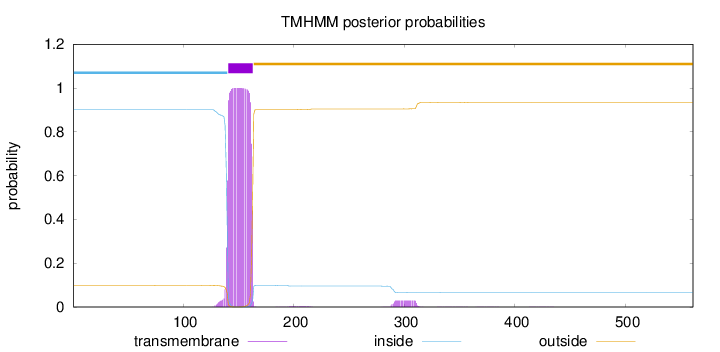
Topology
Length:
561
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.88053
Exp number, first 60 AAs:
0.00061
Total prob of N-in:
0.90107
inside
1 - 140
TMhelix
141 - 163
outside
164 - 561
Population Genetic Test Statistics
Pi
221.33364
Theta
207.086708
Tajima's D
1.548623
CLR
0.13215
CSRT
0.79946002699865
Interpretation
Uncertain
