Gene
KWMTBOMO01837
Pre Gene Modal
BGIBMGA005973
Annotation
PREDICTED:_dedicator_of_cytokinesis_protein_9_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.199
Sequence
CDS
ATGCCTCCGGTCAATTCTAAAGTAAAAAAGGCTGGCGCAGAATATGAGAACATTATCACCCGCAAATGCACAGCACTTGATTTTGAACAATACATTCAAGATAATAAAACTTTACTTCTGAATGATCCATTGAGGGATATCCTCTTATACCCAACAGATGATATTTCAAGTGTAGTCCTTCCTCGGCGCTGGAGAACTGTCACTACTGCAGTTCCAGATGTGAGGATAGCTGCATCGTGTCCCTTGCTCACGAGACAAGCTCTGCTCAGCTATGCCTCTAGTTGGAATCTTATTCACTACAAATATAGCAACTATTCTGGGACTTATGTAAATCTGCCAAGGCTCGTCGTAAGCAAACTGCCTGAAGAGGTATACGACATAGACGCGGAAACGGAAAACGATGAGAACCAACCTACGAAAGTTGACCTCGGTACTAAAGAAGGATATTTGCTCAAAGGCCCTGAGATCGGATCGGATAGGCTCTTCGGAAATCTCGCTTCTAAATCGTTTAAGAAACGCTATTGCTCAATGATCAGAGAAGCTGACGGCACCTACATACTCGAAATTTATAAAGATGATAAGAAAACGGAAACAAAGCTCACTATCGTAATGGATTTTTGCTCTGATGTTGTTCGTAATACCAAACGCGGCAGATTTAGCTTCGAGCTACGCATGTCTGGCGGTAAAGGGTACACGTTTTCAGCGGAAAACGAAGAGGAAATGAATGAATGGATTAACACATTTAAAGCAACGCTAAAGAAAAACCAAGATAATCAGGAAACAAACCAAAGCAACGAGGCATTAGATAAAGGTTAA
Protein
MPPVNSKVKKAGAEYENIITRKCTALDFEQYIQDNKTLLLNDPLRDILLYPTDDISSVVLPRRWRTVTTAVPDVRIAASCPLLTRQALLSYASSWNLIHYKYSNYSGTYVNLPRLVVSKLPEEVYDIDAETENDENQPTKVDLGTKEGYLLKGPEIGSDRLFGNLASKSFKKRYCSMIREADGTYILEIYKDDKKTETKLTIVMDFCSDVVRNTKRGRFSFELRMSGGKGYTFSAENEEEMNEWINTFKATLKKNQDNQETNQSNEALDKG
Summary
Similarity
Belongs to the DOCK family.
Uniprot
H9J8Y1
A0A2H1WGF0
A0A2A4K8U3
A0A2A4K9Q4
A0A3S2M7K6
A0A212F631
+ More
A0A194R342 A0A0L7LP83 A0A194PKT7 A0A2J7R7J7 A0A1B6DJI0 A0A1B6HAI3 A0A1B6ESF3 A0A2J7R7J9 A0A1Y1JXC9 A0A023F2T5 E2BSG6 E0VF86 A0A0L7RKD5 A0A2A3EH22 A0A088AQN5 A0A195BLI2 T1H933 A0A158NLQ7 A0A154NX43 A0A151IAP8 F4X2S7 A0A195FVS9 A0A0J7KRZ1 A0A310SEX8 A0A1W4X9S1 A0A151IU48 A0A151WK12 E2A4X4 A0A3L8DIN0 A0A0C9RH84 A0A0C9R4D2 A0A0C9Q184 E9IW93 U4U1X9 N6TER4 A0A146LPR0 A0A146MIG5 A0A139WGV9 D6WLQ6 A0A1B0D1K7 A0A336MD19 K7J1E7 A0A336LXM8 A0A336LXY2 A0A336M6C0 A0A2H8TJ44 W4VS04 A0A1J1I0R3 A0A232FFG2 J9JVV8 A0A182UJN8 V5IG66 A0A131Y3U6 A0A182RWD5 A0A182YP55 A0A1A9WBW1 A0A182VSH9 A0A1B0C438 A0A1A9X8A9 A0A1A9VA10 A0A1B0AIT5 A0A1B0G6A2 A0A182MBY6 A0A0Q5WAN3 A0A0Q5WBE3 A0A0Q5WNG9 A0A0Q5WK71 B3N6K7 B4NZJ8 A0A0Q5WKD4 A0A0Q5WPE5 A0A0Q5WBG8 A0A0R1DQY6 A0A0R1DJF7
A0A194R342 A0A0L7LP83 A0A194PKT7 A0A2J7R7J7 A0A1B6DJI0 A0A1B6HAI3 A0A1B6ESF3 A0A2J7R7J9 A0A1Y1JXC9 A0A023F2T5 E2BSG6 E0VF86 A0A0L7RKD5 A0A2A3EH22 A0A088AQN5 A0A195BLI2 T1H933 A0A158NLQ7 A0A154NX43 A0A151IAP8 F4X2S7 A0A195FVS9 A0A0J7KRZ1 A0A310SEX8 A0A1W4X9S1 A0A151IU48 A0A151WK12 E2A4X4 A0A3L8DIN0 A0A0C9RH84 A0A0C9R4D2 A0A0C9Q184 E9IW93 U4U1X9 N6TER4 A0A146LPR0 A0A146MIG5 A0A139WGV9 D6WLQ6 A0A1B0D1K7 A0A336MD19 K7J1E7 A0A336LXM8 A0A336LXY2 A0A336M6C0 A0A2H8TJ44 W4VS04 A0A1J1I0R3 A0A232FFG2 J9JVV8 A0A182UJN8 V5IG66 A0A131Y3U6 A0A182RWD5 A0A182YP55 A0A1A9WBW1 A0A182VSH9 A0A1B0C438 A0A1A9X8A9 A0A1A9VA10 A0A1B0AIT5 A0A1B0G6A2 A0A182MBY6 A0A0Q5WAN3 A0A0Q5WBE3 A0A0Q5WNG9 A0A0Q5WK71 B3N6K7 B4NZJ8 A0A0Q5WKD4 A0A0Q5WPE5 A0A0Q5WBG8 A0A0R1DQY6 A0A0R1DJF7
Pubmed
EMBL
BABH01004863
BABH01004864
ODYU01008500
SOQ52159.1
NWSH01000024
PCG80627.1
+ More
PCG80628.1 RSAL01000014 RVE53287.1 AGBW02010086 OWR49184.1 KQ460855 KPJ11939.1 JTDY01000422 KOB77262.1 KQ459601 KPI93942.1 NEVH01006731 PNF36808.1 GEDC01011528 JAS25770.1 GECU01036033 JAS71673.1 GECZ01028883 JAS40886.1 PNF36810.1 GEZM01100429 JAV52871.1 GBBI01003209 JAC15503.1 GL450233 EFN81354.1 AAZO01001761 DS235107 EEB12042.1 KQ414578 KOC71206.1 KZ288253 PBC31020.1 KQ976450 KYM85662.1 ACPB03001105 ACPB03001106 ADTU01019667 KQ434777 KZC04246.1 KQ978186 KYM96573.1 GL888591 EGI59273.1 KQ981219 KYN44412.1 LBMM01003898 KMQ93014.1 KQ769580 OAD52855.1 KQ980968 KYN10954.1 KQ983020 KYQ48202.1 GL436756 EFN71516.1 QOIP01000007 RLU20211.1 GBYB01007665 JAG77432.1 GBYB01007667 JAG77434.1 GBYB01007668 JAG77435.1 GL766458 EFZ15167.1 KB631904 ERL87077.1 APGK01040726 APGK01040727 KB740984 ENN76248.1 GDHC01009897 JAQ08732.1 GDHC01000169 JAQ18460.1 KQ971343 KYB27228.1 EFA03420.2 AJVK01010287 AJVK01010288 AJVK01010289 AJVK01010290 UFQT01000393 SSX23928.1 UFQT01000285 SSX22806.1 SSX22805.1 SSX23927.1 GFXV01002254 MBW14059.1 GANO01000103 JAB59768.1 CVRI01000038 CRK93787.1 NNAY01000330 OXU29169.1 ABLF02028408 ABLF02028415 GANP01007222 JAB77246.1 GEFM01002618 JAP73178.1 JXJN01025268 CCAG010005175 AXCM01002695 CH954177 KQS70695.1 KQS70701.1 KQS70698.1 KQS70699.1 EDV59223.2 CM000157 EDW87747.2 KQS70697.1 KQS70696.1 KQS70700.1 KRJ97369.1 KRJ97372.1
PCG80628.1 RSAL01000014 RVE53287.1 AGBW02010086 OWR49184.1 KQ460855 KPJ11939.1 JTDY01000422 KOB77262.1 KQ459601 KPI93942.1 NEVH01006731 PNF36808.1 GEDC01011528 JAS25770.1 GECU01036033 JAS71673.1 GECZ01028883 JAS40886.1 PNF36810.1 GEZM01100429 JAV52871.1 GBBI01003209 JAC15503.1 GL450233 EFN81354.1 AAZO01001761 DS235107 EEB12042.1 KQ414578 KOC71206.1 KZ288253 PBC31020.1 KQ976450 KYM85662.1 ACPB03001105 ACPB03001106 ADTU01019667 KQ434777 KZC04246.1 KQ978186 KYM96573.1 GL888591 EGI59273.1 KQ981219 KYN44412.1 LBMM01003898 KMQ93014.1 KQ769580 OAD52855.1 KQ980968 KYN10954.1 KQ983020 KYQ48202.1 GL436756 EFN71516.1 QOIP01000007 RLU20211.1 GBYB01007665 JAG77432.1 GBYB01007667 JAG77434.1 GBYB01007668 JAG77435.1 GL766458 EFZ15167.1 KB631904 ERL87077.1 APGK01040726 APGK01040727 KB740984 ENN76248.1 GDHC01009897 JAQ08732.1 GDHC01000169 JAQ18460.1 KQ971343 KYB27228.1 EFA03420.2 AJVK01010287 AJVK01010288 AJVK01010289 AJVK01010290 UFQT01000393 SSX23928.1 UFQT01000285 SSX22806.1 SSX22805.1 SSX23927.1 GFXV01002254 MBW14059.1 GANO01000103 JAB59768.1 CVRI01000038 CRK93787.1 NNAY01000330 OXU29169.1 ABLF02028408 ABLF02028415 GANP01007222 JAB77246.1 GEFM01002618 JAP73178.1 JXJN01025268 CCAG010005175 AXCM01002695 CH954177 KQS70695.1 KQS70701.1 KQS70698.1 KQS70699.1 EDV59223.2 CM000157 EDW87747.2 KQS70697.1 KQS70696.1 KQS70700.1 KRJ97369.1 KRJ97372.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000037510
+ More
UP000053268 UP000235965 UP000008237 UP000009046 UP000053825 UP000242457 UP000005203 UP000078540 UP000015103 UP000005205 UP000076502 UP000078542 UP000007755 UP000078541 UP000036403 UP000192223 UP000078492 UP000075809 UP000000311 UP000279307 UP000030742 UP000019118 UP000007266 UP000092462 UP000002358 UP000183832 UP000215335 UP000007819 UP000075902 UP000075900 UP000076408 UP000091820 UP000075920 UP000092460 UP000092443 UP000078200 UP000092445 UP000092444 UP000075883 UP000008711 UP000002282
UP000053268 UP000235965 UP000008237 UP000009046 UP000053825 UP000242457 UP000005203 UP000078540 UP000015103 UP000005205 UP000076502 UP000078542 UP000007755 UP000078541 UP000036403 UP000192223 UP000078492 UP000075809 UP000000311 UP000279307 UP000030742 UP000019118 UP000007266 UP000092462 UP000002358 UP000183832 UP000215335 UP000007819 UP000075902 UP000075900 UP000076408 UP000091820 UP000075920 UP000092460 UP000092443 UP000078200 UP000092445 UP000092444 UP000075883 UP000008711 UP000002282
PRIDE
Interpro
IPR035892
C2_domain_sf
+ More
IPR027007 DHR-1_domain
IPR026791 DOCK
IPR021816 DOCK_C/D_N
IPR001849 PH_domain
IPR010703 DOCK_C
IPR027357 DHR-2
IPR011993 PH-like_dom_sf
IPR037809 C2_Dock-D
IPR016024 ARM-type_fold
IPR009145 U2AF_small
IPR003954 RRM_dom_euk
IPR001932 PPM-type_phosphatase_dom
IPR035979 RBD_domain_sf
IPR000571 Znf_CCCH
IPR036457 PPM-type_dom_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR027007 DHR-1_domain
IPR026791 DOCK
IPR021816 DOCK_C/D_N
IPR001849 PH_domain
IPR010703 DOCK_C
IPR027357 DHR-2
IPR011993 PH-like_dom_sf
IPR037809 C2_Dock-D
IPR016024 ARM-type_fold
IPR009145 U2AF_small
IPR003954 RRM_dom_euk
IPR001932 PPM-type_phosphatase_dom
IPR035979 RBD_domain_sf
IPR000571 Znf_CCCH
IPR036457 PPM-type_dom_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
Gene 3D
ProteinModelPortal
H9J8Y1
A0A2H1WGF0
A0A2A4K8U3
A0A2A4K9Q4
A0A3S2M7K6
A0A212F631
+ More
A0A194R342 A0A0L7LP83 A0A194PKT7 A0A2J7R7J7 A0A1B6DJI0 A0A1B6HAI3 A0A1B6ESF3 A0A2J7R7J9 A0A1Y1JXC9 A0A023F2T5 E2BSG6 E0VF86 A0A0L7RKD5 A0A2A3EH22 A0A088AQN5 A0A195BLI2 T1H933 A0A158NLQ7 A0A154NX43 A0A151IAP8 F4X2S7 A0A195FVS9 A0A0J7KRZ1 A0A310SEX8 A0A1W4X9S1 A0A151IU48 A0A151WK12 E2A4X4 A0A3L8DIN0 A0A0C9RH84 A0A0C9R4D2 A0A0C9Q184 E9IW93 U4U1X9 N6TER4 A0A146LPR0 A0A146MIG5 A0A139WGV9 D6WLQ6 A0A1B0D1K7 A0A336MD19 K7J1E7 A0A336LXM8 A0A336LXY2 A0A336M6C0 A0A2H8TJ44 W4VS04 A0A1J1I0R3 A0A232FFG2 J9JVV8 A0A182UJN8 V5IG66 A0A131Y3U6 A0A182RWD5 A0A182YP55 A0A1A9WBW1 A0A182VSH9 A0A1B0C438 A0A1A9X8A9 A0A1A9VA10 A0A1B0AIT5 A0A1B0G6A2 A0A182MBY6 A0A0Q5WAN3 A0A0Q5WBE3 A0A0Q5WNG9 A0A0Q5WK71 B3N6K7 B4NZJ8 A0A0Q5WKD4 A0A0Q5WPE5 A0A0Q5WBG8 A0A0R1DQY6 A0A0R1DJF7
A0A194R342 A0A0L7LP83 A0A194PKT7 A0A2J7R7J7 A0A1B6DJI0 A0A1B6HAI3 A0A1B6ESF3 A0A2J7R7J9 A0A1Y1JXC9 A0A023F2T5 E2BSG6 E0VF86 A0A0L7RKD5 A0A2A3EH22 A0A088AQN5 A0A195BLI2 T1H933 A0A158NLQ7 A0A154NX43 A0A151IAP8 F4X2S7 A0A195FVS9 A0A0J7KRZ1 A0A310SEX8 A0A1W4X9S1 A0A151IU48 A0A151WK12 E2A4X4 A0A3L8DIN0 A0A0C9RH84 A0A0C9R4D2 A0A0C9Q184 E9IW93 U4U1X9 N6TER4 A0A146LPR0 A0A146MIG5 A0A139WGV9 D6WLQ6 A0A1B0D1K7 A0A336MD19 K7J1E7 A0A336LXM8 A0A336LXY2 A0A336M6C0 A0A2H8TJ44 W4VS04 A0A1J1I0R3 A0A232FFG2 J9JVV8 A0A182UJN8 V5IG66 A0A131Y3U6 A0A182RWD5 A0A182YP55 A0A1A9WBW1 A0A182VSH9 A0A1B0C438 A0A1A9X8A9 A0A1A9VA10 A0A1B0AIT5 A0A1B0G6A2 A0A182MBY6 A0A0Q5WAN3 A0A0Q5WBE3 A0A0Q5WNG9 A0A0Q5WK71 B3N6K7 B4NZJ8 A0A0Q5WKD4 A0A0Q5WPE5 A0A0Q5WBG8 A0A0R1DQY6 A0A0R1DJF7
PDB
1WG7
E-value=2.26554e-14,
Score=190
Ontologies
PANTHER
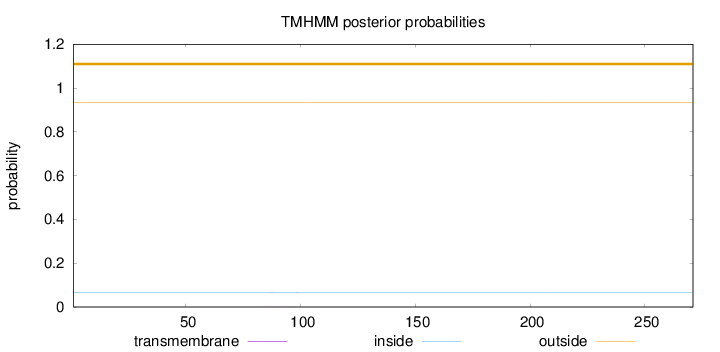
Topology
Length:
271
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.004
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06712
outside
1 - 271
Population Genetic Test Statistics
Pi
299.758763
Theta
199.823546
Tajima's D
1.320195
CLR
0.383908
CSRT
0.746362681865907
Interpretation
Uncertain
