Gene
KWMTBOMO01836
Pre Gene Modal
BGIBMGA005973
Annotation
PREDICTED:_dedicator_of_cytokinesis_protein_9_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.306
Sequence
CDS
ATGAACCCCCTCCTAATCCGGTACTCCCGGGAGACGGACCTGTCTATAGCGGCGGCGCGCAACGAGAGCCGCTTCAACGTGTTCATGTTGCCGTACAAACGAGCGCCGTCGCCAGAACCTCAGTTGGAACCATTCAAAGAACATTTTGGCCAAAGAATATTGCTCAAATGCGAAAGTTTAAAGTTTAGGTTACAAGCGCCCATAGACGGGGACAAAGAGTTGTTATGTCAGGTCGAACCTTACTACACTCGTCTCGCGTTGTACGACGCGAGATGCGGCAAGAAACTAACCGAGGACTTCCACTTCGATCTGAACCACGACGCCGTCAAGGATCTAGTCAAGGACGACGAGTGCACAAAGTCATTAGTCGAAAACTTTAGTTACGATGTTAAATTAGATGTAAAACAGATACCGGAGGAATGGTATCGGTCGAAGAGACAGGTGGTTTTATCGGTGAATAATCCACATCCGGACTTATTTTTAGTAGTTAAAATAGACAAAATACTCCAAGGTCATGTTAGTCAAGTTTTGGAACCGTATATGAAAGCGACCAAGGACCCCAGGCTCGGTCTTAAGGTTCACAAAAACGTGCAAGCGTATGGGAATCGCGTCGGAAACTACAAAATGCCCTTCGCTTGGGCAGCAAAGCCTTTGTACAAAATGTACAGCAATGATTTGGATGTAACCTCAGAGTTTTCTGCGATTTACCGACAGGAGCCTAATAAGATGTCCGATGATGACTTATTAAAAATTTTATCTGATTATCGGAAACCCGAGAAGCTGGCTCGACTAACCGTTATACCCGGCTGGCTCTCTATAAATATACAACAGACAAGCGAACAAATACCAAATTGTATGACGAGTGCGTACGCGGCCCTTAAGCCGTTTCCGCTGCCGCCGGCTGCGCCTCTGACGCTAGAGCTCGCAACACTGAACGTGGAGCCGGAGCAACCGTACGCGGCCTACGTGCATCACCTCTACGTGCGACCGCTTGCTCTGAATTTCGAATCTCAAAAAATGTTCGCCAGGGCTAGGAACATCGCATGCACTATCGAACTCAGGAATAGCGACACGGGCGATAATGAACCGCTACAGGCAATATATGGCCGCACAGGAATGACGAGTCAATACAGATGTTGCGTTTTACATCATAACGCTAATCCGGCCTGGTCGGACGAAGCGAAAATCCGTCTGCCCGCCACAATTAATTCACAGCATCATTTACTATTCACATTCTACCACATCTCTTGCGATATCGCCAAGAAAAACGACACGAATGTCGAAACGTGTATCGGGTACGCGTGGGTCCCGCTATTGAAAAACGATAAATTTGTCGATGAGCTTATAAACCTACCGGTTTCCACACATTTGCCGTCTGGATATTTATCGATACAACCTCTCGGGCTCGGCAAAGGGAATTCCGGACCGGAAATAGTATGGATCGAAGGAGAGAAGCAACTGTTCCGGTGTCATATAGTACTAGATTCGACAGTCGCAACCAGGGATAATCACTTGCATAATCTGTTCAATCATGCGGAGAGATTGGTGAAGAGTAACTCGCCTCCCACGTCGCCGTCTCCTCCGCCATGGAATGACGTCTGCAACGCACTGAAAGCCGCACACGCAGTCAAACTCAGCTCACTTGTTGGTTTCTTACCGACAATCCTAAACCAGCTGTTCGAATTGATGACTGTTGAAAAGAGTTACACATCGGAAATGGGCTATCAGATTGTCAAATTAATTGTTCACTTCGTTCATTTGATTCATGATTACGGGAGAAAAGATTTGTTGGACAGCTATGTGAAGTATGTTTTTAACTGTCTTGAATATAAGTTGCACACAGTTCTAACCGCGCCTCTACATATGTTTGTGGACCCCAATCAACAAGATTTTTTGCTAAATCATAAATTCATGCAGTACTCCAACTTCTTCTTCGAAATCATTGTGAAGAGTATGGCTCAGTATTTAATTAACACAGGACGAATAAAGATGTCCAGGAATGAGAGGTTTCATTCAGATTTATTAGAGAATATTGACAAACTGGTCACGATGGTAGAGCCGTCGTACATTCTACAACAGCCGATGCAGACTCATATTTTTAATAAAAATCTTGCCATTTTTCTTAAGTCTTGCTTATCGTTCATGGATCGAGGATTTGTATTTCGTCAAATAAAGAAATACCTCGACAAGTTTAAGGCGTGCGATCCCAAAGCTCTGTTCGACTTCAAATTTACATTCTTACAGACAATTTGTTTGCACGAACATTACGTACCCTTCAATTTACCTCTTCAAGCGAATAAGATCGGTAAAGACTGTGATACTGATCCTGCTAAACTAAAATTAACGGAAGACTTCATCATGCGACATTTTCTAGCCGGAACACTACTGAAACATGTGGAACAGTCACTTCGTGAAGCACCTCAGAAACGCCGAGTGGCTTTGAACGTGCTCAGAACGTTGCTGACGAAACACGAACACGATGATCGATATCGATCAAAGCAGTGCCGTGCGAGGATAGCACATCTCTATTCGCCCTGGCTTACTATTGTTTTAGATAACGCTCATCGCCTCGTCACCAAGTCTCTAGCCAGTCCAGTGCTAGAGAACGGGGTAGACAAGGTGGACGGCGACGCTACCAACACCACTGCCAACTGCGCGAAGGAAGCCGCTTCAGTCAACAGTACCCCGCGCAGAAATAGATTGACTTTGCATTTCGAGCACACGCCGGCTCGCAATTCTACTCACTTCAAAGAACCATCAAACGTTCAAAGTAATCACTTGTATGCAAAAGATCCCGGCAACAACGCATCGCACAGTTCCCTTGAATCGGTTTCGACCATGTCCGGCGGCGACTCGCTTCCTCGGAATATAGGCAAGTTAGATTTGAGCGAAATTGGTGATCAAGTTAATCGTTTCGGCGTGATGTCTGCCGAGGAAGTCCGCGATGTTCTGTTATGCTTCCTATTTGTATTGAAATATTTGGATGAAGAGAGACTCGTTGATTGGTGGAAGACGCTATCTGAGGCCTACATACAGTCATTCTTTAACGTTTTAGAGATCTGCGCAGAACAGTTTCAGTATGTTGGAAGAAAAAAGATTCTGAGTGAACTGAAGGCGACAACGCCGGACTGCAACGGCAAGCCGAAGCCAGTGAAGGCCAGGACGCTGCCCGCCCGGATGAGCCCACCGGATTTTTCCAAGGAACCTCCCATAATTGTCGAGAAGAACAACACAGTTAACAGAGAAAATCTGGCTAATACTGCCACAATTACAACAGAATTAGAAGCGATGGCGGAACACACGGTGTTAGCAGCAGGATGCTTGTCCACAGAAGTGGGCCTCATTATAATAGATAGATCGTGCTTATTCATGAAAAATCTCGGAGCGGCTAATGGAGTTAGCGCCAAACCGTATCTAAAACTACTACAGTGTCCACAAAGCGAAACTCTGTACAAACACCTGTTCGCAGCCTTGCGAGCCTATATCAACCAGTTCTCTGAAACACTATTCGAAGGGGGCAGCACGTTGTGCGCCGGCGTGGTGTCGGGCGTGGTGCGGCTGTGCGAGGCGCGCGCGGCGTGGCTGCGGCGGGAGGCAGCGGCCGCCCTCTATCTGCTGATGCGGGCTAACTTCCAACACGGAGCCGGTGGGGATCTCACCAGAGTTCATTTGCAAGTCATAATGGCGGTCTCAAAACTGTTAGACAGTTCAAACGCACTAAACTGCAAACGTTTCCAAGAGAGTCTGTCCGTTATAAACTGCTTCGCTACAGGTGACAAGGCAATGAAAGGAACGGGATTGTCGAGCGAAGTGGCGGAGCTGATGCGGCGCGTCCGTACAGTAGTGGGCGCGCGGGCTCGCCTCGCCGGAGTGGGGGGCTCCGGTCTCTCGGGTCTCTCAGGTGCCGGTAGCGTCCACCTCGCGGAGCTACAGCACGCTCTGGCGGCGTCGTGCCGCGCCACGCCGCGCCTGCGCCACGCCTGGCTCGCGACGCTCGCGGAGCACAACGCGCGACAAGGAGACCACGATGAGGCCATGTGCTGTCAGTTGCACATAGCGGCCCTCATCGCCGAGTACTTGAAGCTGCGCGGCGCCATACAGTGGGGGGCCGATGTCTTCGAAGACATTTCCAAGAACATTCCACGCGAAGAGACCGGCCTCAAGATAGACGAAGGTAAATAA
Protein
MNPLLIRYSRETDLSIAAARNESRFNVFMLPYKRAPSPEPQLEPFKEHFGQRILLKCESLKFRLQAPIDGDKELLCQVEPYYTRLALYDARCGKKLTEDFHFDLNHDAVKDLVKDDECTKSLVENFSYDVKLDVKQIPEEWYRSKRQVVLSVNNPHPDLFLVVKIDKILQGHVSQVLEPYMKATKDPRLGLKVHKNVQAYGNRVGNYKMPFAWAAKPLYKMYSNDLDVTSEFSAIYRQEPNKMSDDDLLKILSDYRKPEKLARLTVIPGWLSINIQQTSEQIPNCMTSAYAALKPFPLPPAAPLTLELATLNVEPEQPYAAYVHHLYVRPLALNFESQKMFARARNIACTIELRNSDTGDNEPLQAIYGRTGMTSQYRCCVLHHNANPAWSDEAKIRLPATINSQHHLLFTFYHISCDIAKKNDTNVETCIGYAWVPLLKNDKFVDELINLPVSTHLPSGYLSIQPLGLGKGNSGPEIVWIEGEKQLFRCHIVLDSTVATRDNHLHNLFNHAERLVKSNSPPTSPSPPPWNDVCNALKAAHAVKLSSLVGFLPTILNQLFELMTVEKSYTSEMGYQIVKLIVHFVHLIHDYGRKDLLDSYVKYVFNCLEYKLHTVLTAPLHMFVDPNQQDFLLNHKFMQYSNFFFEIIVKSMAQYLINTGRIKMSRNERFHSDLLENIDKLVTMVEPSYILQQPMQTHIFNKNLAIFLKSCLSFMDRGFVFRQIKKYLDKFKACDPKALFDFKFTFLQTICLHEHYVPFNLPLQANKIGKDCDTDPAKLKLTEDFIMRHFLAGTLLKHVEQSLREAPQKRRVALNVLRTLLTKHEHDDRYRSKQCRARIAHLYSPWLTIVLDNAHRLVTKSLASPVLENGVDKVDGDATNTTANCAKEAASVNSTPRRNRLTLHFEHTPARNSTHFKEPSNVQSNHLYAKDPGNNASHSSLESVSTMSGGDSLPRNIGKLDLSEIGDQVNRFGVMSAEEVRDVLLCFLFVLKYLDEERLVDWWKTLSEAYIQSFFNVLEICAEQFQYVGRKKILSELKATTPDCNGKPKPVKARTLPARMSPPDFSKEPPIIVEKNNTVNRENLANTATITTELEAMAEHTVLAAGCLSTEVGLIIIDRSCLFMKNLGAANGVSAKPYLKLLQCPQSETLYKHLFAALRAYINQFSETLFEGGSTLCAGVVSGVVRLCEARAAWLRREAAAALYLLMRANFQHGAGGDLTRVHLQVIMAVSKLLDSSNALNCKRFQESLSVINCFATGDKAMKGTGLSSEVAELMRRVRTVVGARARLAGVGGSGLSGLSGAGSVHLAELQHALAASCRATPRLRHAWLATLAEHNARQGDHDEAMCCQLHIAALIAEYLKLRGAIQWGADVFEDISKNIPREETGLKIDEGK
Summary
Similarity
Belongs to the DOCK family.
Uniprot
A0A2A4K8U3
A0A2A4K9Q4
H9J8Y1
A0A2H1WGF0
A0A3S2M7K6
A0A194R342
+ More
A0A212F631 E9IW93 A0A154NX43 A0A158NLQ7 A0A195FVS9 F4X2S7 E2A4X4 A0A088AQN5 A0A151IU48 A0A2A3EH22 E2BSG6 A0A026WF93 A0A0L7RKD5 A0A195BLI2 A0A151WK12 A0A3L8DIN0 A0A151IAP8 A0A067RL42 A0A2J7R7J9 D6WLQ6 A0A139WGV9 A0A310SEX8 N6TER4 A0A1Y1JU94 U4U1X9 A0A1Y1JXC9 A0A0C9Q184 E0VF86 A0A1B6I8Z2 A0A0C9RH84 A0A0C9R4D2 A0A1B6I6N9 A0A1W4X9S1 A0A1B0D1K7 A0A232FFG2 K7J1E7 A0A023F2T5 A0A182FNB8 A0A1I8PQ35 A0A1I8PPW5 A0A1I8PQ16 A0A1I8PQ33 A0A1I8PPX8 A0A1I8PPX3 A0A1I8PQ27 A0A1I8PPX0 A0A1I8PQ24 A0A182JHE5 A0A0L0CGE9 A0A182VSH9 A0A182P4N5 A0A1J1I0R3 W8B9Y8 W8AEL3 W8B0C6 B4JQF8 A0A453Z3H1 A0A182V3Y8 A0A182RWD5 A0A182YP55 A0NF07 A0A182K3W5 A0A182MBY6 A0A182NT68 A0A0Q9X647 A0A0Q9X591 A0A1S4GZB0 A0A0Q9XGS4 A0A0Q9X599 A0A182L027 A0A453YJT6 A0A0Q9XI10 A0A0A1WGY8 A0A0Q9XIB8 A0A0Q9XFC8 A0A0Q9XEV2 A0A0Q9X5A8 A0A0Q9XEA3 A0A0Q9XGI5 B4KKA1 A0A182Q331 A0A0Q9WAR9 A0A0Q9WBF9 A0A1B0C438 A0A0Q9WN78 A0A0Q9WAS2 A0A1A9X8A9 A0A1B0G6A2 A0A0Q9WAU6 A0A0Q9WLP5 A0A0Q9WKW6 A0A0Q9WLV6 B4LRV0
A0A212F631 E9IW93 A0A154NX43 A0A158NLQ7 A0A195FVS9 F4X2S7 E2A4X4 A0A088AQN5 A0A151IU48 A0A2A3EH22 E2BSG6 A0A026WF93 A0A0L7RKD5 A0A195BLI2 A0A151WK12 A0A3L8DIN0 A0A151IAP8 A0A067RL42 A0A2J7R7J9 D6WLQ6 A0A139WGV9 A0A310SEX8 N6TER4 A0A1Y1JU94 U4U1X9 A0A1Y1JXC9 A0A0C9Q184 E0VF86 A0A1B6I8Z2 A0A0C9RH84 A0A0C9R4D2 A0A1B6I6N9 A0A1W4X9S1 A0A1B0D1K7 A0A232FFG2 K7J1E7 A0A023F2T5 A0A182FNB8 A0A1I8PQ35 A0A1I8PPW5 A0A1I8PQ16 A0A1I8PQ33 A0A1I8PPX8 A0A1I8PPX3 A0A1I8PQ27 A0A1I8PPX0 A0A1I8PQ24 A0A182JHE5 A0A0L0CGE9 A0A182VSH9 A0A182P4N5 A0A1J1I0R3 W8B9Y8 W8AEL3 W8B0C6 B4JQF8 A0A453Z3H1 A0A182V3Y8 A0A182RWD5 A0A182YP55 A0NF07 A0A182K3W5 A0A182MBY6 A0A182NT68 A0A0Q9X647 A0A0Q9X591 A0A1S4GZB0 A0A0Q9XGS4 A0A0Q9X599 A0A182L027 A0A453YJT6 A0A0Q9XI10 A0A0A1WGY8 A0A0Q9XIB8 A0A0Q9XFC8 A0A0Q9XEV2 A0A0Q9X5A8 A0A0Q9XEA3 A0A0Q9XGI5 B4KKA1 A0A182Q331 A0A0Q9WAR9 A0A0Q9WBF9 A0A1B0C438 A0A0Q9WN78 A0A0Q9WAS2 A0A1A9X8A9 A0A1B0G6A2 A0A0Q9WAU6 A0A0Q9WLP5 A0A0Q9WKW6 A0A0Q9WLV6 B4LRV0
Pubmed
EMBL
NWSH01000024
PCG80627.1
PCG80628.1
BABH01004863
BABH01004864
ODYU01008500
+ More
SOQ52159.1 RSAL01000014 RVE53287.1 KQ460855 KPJ11939.1 AGBW02010086 OWR49184.1 GL766458 EFZ15167.1 KQ434777 KZC04246.1 ADTU01019667 KQ981219 KYN44412.1 GL888591 EGI59273.1 GL436756 EFN71516.1 KQ980968 KYN10954.1 KZ288253 PBC31020.1 GL450233 EFN81354.1 KK107238 EZA54722.1 KQ414578 KOC71206.1 KQ976450 KYM85662.1 KQ983020 KYQ48202.1 QOIP01000007 RLU20211.1 KQ978186 KYM96573.1 KK852561 KDR21325.1 NEVH01006731 PNF36810.1 KQ971343 EFA03420.2 KYB27228.1 KQ769580 OAD52855.1 APGK01040726 APGK01040727 KB740984 ENN76248.1 GEZM01100428 JAV52872.1 KB631904 ERL87077.1 GEZM01100429 JAV52871.1 GBYB01007668 JAG77435.1 AAZO01001761 DS235107 EEB12042.1 GECU01024301 JAS83405.1 GBYB01007665 JAG77432.1 GBYB01007667 JAG77434.1 GECU01025115 JAS82591.1 AJVK01010287 AJVK01010288 AJVK01010289 AJVK01010290 NNAY01000330 OXU29169.1 GBBI01003209 JAC15503.1 JRES01000438 KNC31320.1 CVRI01000038 CRK93787.1 GAMC01019911 JAB86644.1 GAMC01019910 JAB86645.1 GAMC01019909 JAB86646.1 CH916372 EDV99138.1 AAAB01008964 EAU76295.2 AXCM01002695 CH933807 KRG03333.1 KRG03334.1 KRG03335.1 KRG03327.1 APCN01000736 KRG03336.1 GBXI01016023 JAC98268.1 KRG03331.1 KRG03326.1 KRG03330.1 KRG03328.1 KRG03332.1 KRG03329.1 EDW12632.2 AXCN02000728 CH940649 KRF81801.1 KRF81810.1 JXJN01025268 KRF81808.1 KRF81805.1 CCAG010005175 KRF81800.1 KRF81806.1 KRF81807.1 KRF81803.1 EDW64702.2
SOQ52159.1 RSAL01000014 RVE53287.1 KQ460855 KPJ11939.1 AGBW02010086 OWR49184.1 GL766458 EFZ15167.1 KQ434777 KZC04246.1 ADTU01019667 KQ981219 KYN44412.1 GL888591 EGI59273.1 GL436756 EFN71516.1 KQ980968 KYN10954.1 KZ288253 PBC31020.1 GL450233 EFN81354.1 KK107238 EZA54722.1 KQ414578 KOC71206.1 KQ976450 KYM85662.1 KQ983020 KYQ48202.1 QOIP01000007 RLU20211.1 KQ978186 KYM96573.1 KK852561 KDR21325.1 NEVH01006731 PNF36810.1 KQ971343 EFA03420.2 KYB27228.1 KQ769580 OAD52855.1 APGK01040726 APGK01040727 KB740984 ENN76248.1 GEZM01100428 JAV52872.1 KB631904 ERL87077.1 GEZM01100429 JAV52871.1 GBYB01007668 JAG77435.1 AAZO01001761 DS235107 EEB12042.1 GECU01024301 JAS83405.1 GBYB01007665 JAG77432.1 GBYB01007667 JAG77434.1 GECU01025115 JAS82591.1 AJVK01010287 AJVK01010288 AJVK01010289 AJVK01010290 NNAY01000330 OXU29169.1 GBBI01003209 JAC15503.1 JRES01000438 KNC31320.1 CVRI01000038 CRK93787.1 GAMC01019911 JAB86644.1 GAMC01019910 JAB86645.1 GAMC01019909 JAB86646.1 CH916372 EDV99138.1 AAAB01008964 EAU76295.2 AXCM01002695 CH933807 KRG03333.1 KRG03334.1 KRG03335.1 KRG03327.1 APCN01000736 KRG03336.1 GBXI01016023 JAC98268.1 KRG03331.1 KRG03326.1 KRG03330.1 KRG03328.1 KRG03332.1 KRG03329.1 EDW12632.2 AXCN02000728 CH940649 KRF81801.1 KRF81810.1 JXJN01025268 KRF81808.1 KRF81805.1 CCAG010005175 KRF81800.1 KRF81806.1 KRF81807.1 KRF81803.1 EDW64702.2
Proteomes
UP000218220
UP000005204
UP000283053
UP000053240
UP000007151
UP000076502
+ More
UP000005205 UP000078541 UP000007755 UP000000311 UP000005203 UP000078492 UP000242457 UP000008237 UP000053097 UP000053825 UP000078540 UP000075809 UP000279307 UP000078542 UP000027135 UP000235965 UP000007266 UP000019118 UP000030742 UP000009046 UP000192223 UP000092462 UP000215335 UP000002358 UP000069272 UP000095300 UP000075880 UP000037069 UP000075920 UP000075885 UP000183832 UP000001070 UP000076407 UP000075903 UP000075900 UP000076408 UP000007062 UP000075881 UP000075883 UP000075884 UP000009192 UP000075882 UP000075840 UP000075886 UP000008792 UP000092460 UP000092443 UP000092444
UP000005205 UP000078541 UP000007755 UP000000311 UP000005203 UP000078492 UP000242457 UP000008237 UP000053097 UP000053825 UP000078540 UP000075809 UP000279307 UP000078542 UP000027135 UP000235965 UP000007266 UP000019118 UP000030742 UP000009046 UP000192223 UP000092462 UP000215335 UP000002358 UP000069272 UP000095300 UP000075880 UP000037069 UP000075920 UP000075885 UP000183832 UP000001070 UP000076407 UP000075903 UP000075900 UP000076408 UP000007062 UP000075881 UP000075883 UP000075884 UP000009192 UP000075882 UP000075840 UP000075886 UP000008792 UP000092460 UP000092443 UP000092444
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2A4K8U3
A0A2A4K9Q4
H9J8Y1
A0A2H1WGF0
A0A3S2M7K6
A0A194R342
+ More
A0A212F631 E9IW93 A0A154NX43 A0A158NLQ7 A0A195FVS9 F4X2S7 E2A4X4 A0A088AQN5 A0A151IU48 A0A2A3EH22 E2BSG6 A0A026WF93 A0A0L7RKD5 A0A195BLI2 A0A151WK12 A0A3L8DIN0 A0A151IAP8 A0A067RL42 A0A2J7R7J9 D6WLQ6 A0A139WGV9 A0A310SEX8 N6TER4 A0A1Y1JU94 U4U1X9 A0A1Y1JXC9 A0A0C9Q184 E0VF86 A0A1B6I8Z2 A0A0C9RH84 A0A0C9R4D2 A0A1B6I6N9 A0A1W4X9S1 A0A1B0D1K7 A0A232FFG2 K7J1E7 A0A023F2T5 A0A182FNB8 A0A1I8PQ35 A0A1I8PPW5 A0A1I8PQ16 A0A1I8PQ33 A0A1I8PPX8 A0A1I8PPX3 A0A1I8PQ27 A0A1I8PPX0 A0A1I8PQ24 A0A182JHE5 A0A0L0CGE9 A0A182VSH9 A0A182P4N5 A0A1J1I0R3 W8B9Y8 W8AEL3 W8B0C6 B4JQF8 A0A453Z3H1 A0A182V3Y8 A0A182RWD5 A0A182YP55 A0NF07 A0A182K3W5 A0A182MBY6 A0A182NT68 A0A0Q9X647 A0A0Q9X591 A0A1S4GZB0 A0A0Q9XGS4 A0A0Q9X599 A0A182L027 A0A453YJT6 A0A0Q9XI10 A0A0A1WGY8 A0A0Q9XIB8 A0A0Q9XFC8 A0A0Q9XEV2 A0A0Q9X5A8 A0A0Q9XEA3 A0A0Q9XGI5 B4KKA1 A0A182Q331 A0A0Q9WAR9 A0A0Q9WBF9 A0A1B0C438 A0A0Q9WN78 A0A0Q9WAS2 A0A1A9X8A9 A0A1B0G6A2 A0A0Q9WAU6 A0A0Q9WLP5 A0A0Q9WKW6 A0A0Q9WLV6 B4LRV0
A0A212F631 E9IW93 A0A154NX43 A0A158NLQ7 A0A195FVS9 F4X2S7 E2A4X4 A0A088AQN5 A0A151IU48 A0A2A3EH22 E2BSG6 A0A026WF93 A0A0L7RKD5 A0A195BLI2 A0A151WK12 A0A3L8DIN0 A0A151IAP8 A0A067RL42 A0A2J7R7J9 D6WLQ6 A0A139WGV9 A0A310SEX8 N6TER4 A0A1Y1JU94 U4U1X9 A0A1Y1JXC9 A0A0C9Q184 E0VF86 A0A1B6I8Z2 A0A0C9RH84 A0A0C9R4D2 A0A1B6I6N9 A0A1W4X9S1 A0A1B0D1K7 A0A232FFG2 K7J1E7 A0A023F2T5 A0A182FNB8 A0A1I8PQ35 A0A1I8PPW5 A0A1I8PQ16 A0A1I8PQ33 A0A1I8PPX8 A0A1I8PPX3 A0A1I8PQ27 A0A1I8PPX0 A0A1I8PQ24 A0A182JHE5 A0A0L0CGE9 A0A182VSH9 A0A182P4N5 A0A1J1I0R3 W8B9Y8 W8AEL3 W8B0C6 B4JQF8 A0A453Z3H1 A0A182V3Y8 A0A182RWD5 A0A182YP55 A0NF07 A0A182K3W5 A0A182MBY6 A0A182NT68 A0A0Q9X647 A0A0Q9X591 A0A1S4GZB0 A0A0Q9XGS4 A0A0Q9X599 A0A182L027 A0A453YJT6 A0A0Q9XI10 A0A0A1WGY8 A0A0Q9XIB8 A0A0Q9XFC8 A0A0Q9XEV2 A0A0Q9X5A8 A0A0Q9XEA3 A0A0Q9XGI5 B4KKA1 A0A182Q331 A0A0Q9WAR9 A0A0Q9WBF9 A0A1B0C438 A0A0Q9WN78 A0A0Q9WAS2 A0A1A9X8A9 A0A1B0G6A2 A0A0Q9WAU6 A0A0Q9WLP5 A0A0Q9WKW6 A0A0Q9WLV6 B4LRV0
PDB
2WMO
E-value=1.1103e-07,
Score=139
Ontologies
PANTHER
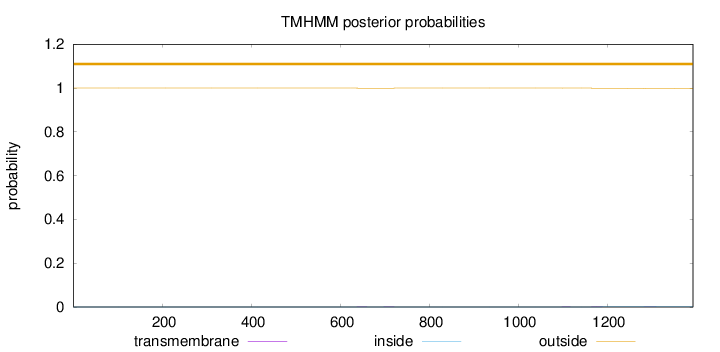
Topology
Length:
1393
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0709400000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 1393
Population Genetic Test Statistics
Pi
263.100566
Theta
160.307914
Tajima's D
1.010335
CLR
1.049235
CSRT
0.663666816659167
Interpretation
Uncertain
