Gene
KWMTBOMO01835
Annotation
PREDICTED:_venom_acid_phosphatase_Acph-1-like_isoform_X1_[Bombyx_mori]
Full name
Venom acid phosphatase Acph-1
Location in the cell
Lysosomal Reliability : 1.342
Sequence
CDS
ATGTCATGTGTAGATAAGTACTGTGGTATAATCGATTGGGCAAAGTCAATCATCATAGGTCGCAGCGACCGTGAATCTCCCACACAATTGGAATACAATGTAGAGGTGCCGACTCGGCGTAGAAAAATGAAAGAGGGCGGTCGCGAGGAGGAGCCGACCAACGCGCCGGAAGCTACACCGGCACCGGGTATGCGCACCGAACACACTTGCTGTTGCCTCGGCGTGTTGGTGGGCATCTCGCTCATAGCCGCCCTCGTAGCAGTAATACTGATGAAGGATACTACGGTAGAAGTCACAATACTTCGACAGGTACATGTGTTGATGTCTCATGGCGAGAGGACTCCAAGTGAACGCGAACTCGAAATGTTAGGAACAAGTCCTCCAGAGCATGTGTTTTCACCTTACGGCGCTGGTGCTATGACAAATGAAGGAAAAATGTTGACTTTCGAAATGGGCGCATTACTTAGAAAGAGATATAACGATTTCTTAGGACCACTTTACGACGCAGAAAGCAGTATCGTTATAGCCTCAGATACAAATTTAAGTAAGATGACAGCTCTGCTGATATCGGCTGGATTGTGGCCGCCGCCGGAAGAGCAGCTCTGGAATGACACAATCGTGTGGCAGCCGGTACCTTACAAATATCCGCCACAAAGCAATGATTTTCTTCTGTACGAAAAAAATTGTCCACGTTATAAACAAGAAAAAGGAAGAATATTGAATGCGTTCGTCGATGAAGGCGTGCTGGTGCCGTATCGGGATCTGTTTCATAAAATTGCGCAAATGACGAACACGAATTTTAGCACACCACAAGAGGCTTATTTCTTAAACAACTTATTTCTTATACAGGACGATATCAAAGTGGCAAATCCGAAATGGGCAAAACACGTTAAAAGGAAATTGATGGACATCGCCCGTCTTGAATACTCAATGATGTTTCACAACAATCTTTTAAGGAAATTGAGTGGCGGGGCTTTATTACATCAAATAATAAAGGAAGCCGTTTCGAACGCATTGGGAGCATCGCGGTTACGCATGATCGTACGCACGGGCACGCCCGTATCCGTGGCAGCCGTGCTGGCGGCGTGCGTGGCGCCGCCGCCGCGTCTCCCCGACCCCGGGGCCGCGTTACTATTCGAACTACACGAACGACAACCGAATAAAGGAAAAAACGAACCATCACTCAACGACGAGCAACGTTTCGGATTTAAGATCTATTACTGGGACGATGATTCAGCTGAACCTCGTCTCATGGAAGTGCCCAATTGCAATTCCTTCTGCACGATTGAACAATTCAAAGAGTTAACGAAATACATAATTGCCCATAATTATGAAGAAGACTGTAAGCTCGTTGCGGAACAAACCAATTGA
Protein
MSCVDKYCGIIDWAKSIIIGRSDRESPTQLEYNVEVPTRRRKMKEGGREEEPTNAPEATPAPGMRTEHTCCCLGVLVGISLIAALVAVILMKDTTVEVTILRQVHVLMSHGERTPSERELEMLGTSPPEHVFSPYGAGAMTNEGKMLTFEMGALLRKRYNDFLGPLYDAESSIVIASDTNLSKMTALLISAGLWPPPEEQLWNDTIVWQPVPYKYPPQSNDFLLYEKNCPRYKQEKGRILNAFVDEGVLVPYRDLFHKIAQMTNTNFSTPQEAYFLNNLFLIQDDIKVANPKWAKHVKRKLMDIARLEYSMMFHNNLLRKLSGGALLHQIIKEAVSNALGASRLRMIVRTGTPVSVAAVLAACVAPPPRLPDPGAALLFELHERQPNKGKNEPSLNDEQRFGFKIYYWDDDSAEPRLMEVPNCNSFCTIEQFKELTKYIIAHNYEEDCKLVAEQTN
Summary
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
Similarity
Belongs to the histidine acid phosphatase family.
Keywords
Allergen
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Reference proteome
Secreted
Signal
Feature
chain Venom acid phosphatase Acph-1
Uniprot
A0A2A4K8T7
A0A2H1WGF6
A0A212F617
A0A3S2NZR1
A0A194R2C4
A0A194PLS5
+ More
H9J9I3 A0A194R7T8 A0A0L7QWI3 A0A067QNZ9 A0A194PS41 A0A2A4K986 A0A212F630 A0A0K8TLP0 K7IZV4 D2A1N0 D2A1N1 D6WAI2 J9K623 Q0PWU9 U4U626 A0A3S2LGG3 A0A1W6EVT0 A0A1S3D5K3 J3JZ60 A0A1S3D5S2 E2C1V8 A0A2S2QXE2 A0A0A9XGP6 A0A146MAA4 F4WFK0 A0A0T6B3P5 A0A3L8DX27 A0A158NSL2 A0A151HZE5 A0A1Y1N416 A0A1L8DYV9 K7IZV5 E2ARU9 E2B168 A0A1B6F055 D3TNZ8 A0A1B6MG81 A0A2A3E9C8 D2A1N2 A0A310SKW5 A0A1L8DZ06 A0A2R4SV17 A0A1Q3EW11 A0A1Q3EW78 A0A1J1I6C6 A0A195FSS6 E9JD78 C1IC99 A0A1A9ZSA2 A0A1Y1N7Z4 A0A1A9WGA5 K7IQ61 A0A0C9R287 A0A1B6JWK6 Q5BLY5 A0A1Q3EVW7 D6WAI1 A0A1Q3EVW8 A0A336L3V8 A0A087ZRC7 A0A1B0GD38 A0A2A3E9H3 A0A023EQR2 N6SVB5 E2C1V9 A0A1B0BCK4 A0A026WAD8 A0A0L7QKE0 A0A088AJL8 A0A232EEI8 A0A154PQI5 A0A2P8YES0 U4UM22 A0A154PK09 A0A060GL20 A0A0M9AAI3 K7IQ63 J3JUU6 A0A170XK82 A0A0L7RCP2 A0A170XK51 E2ABY7 A0A1Y1LK10 A0A1W6EVW2 A0A1W7R8D5 A0A0C9S0C4 A0A0J7KGP1 A0A154P094 A0A154P3P1 A0A310SN00 A0A0C9RY44
H9J9I3 A0A194R7T8 A0A0L7QWI3 A0A067QNZ9 A0A194PS41 A0A2A4K986 A0A212F630 A0A0K8TLP0 K7IZV4 D2A1N0 D2A1N1 D6WAI2 J9K623 Q0PWU9 U4U626 A0A3S2LGG3 A0A1W6EVT0 A0A1S3D5K3 J3JZ60 A0A1S3D5S2 E2C1V8 A0A2S2QXE2 A0A0A9XGP6 A0A146MAA4 F4WFK0 A0A0T6B3P5 A0A3L8DX27 A0A158NSL2 A0A151HZE5 A0A1Y1N416 A0A1L8DYV9 K7IZV5 E2ARU9 E2B168 A0A1B6F055 D3TNZ8 A0A1B6MG81 A0A2A3E9C8 D2A1N2 A0A310SKW5 A0A1L8DZ06 A0A2R4SV17 A0A1Q3EW11 A0A1Q3EW78 A0A1J1I6C6 A0A195FSS6 E9JD78 C1IC99 A0A1A9ZSA2 A0A1Y1N7Z4 A0A1A9WGA5 K7IQ61 A0A0C9R287 A0A1B6JWK6 Q5BLY5 A0A1Q3EVW7 D6WAI1 A0A1Q3EVW8 A0A336L3V8 A0A087ZRC7 A0A1B0GD38 A0A2A3E9H3 A0A023EQR2 N6SVB5 E2C1V9 A0A1B0BCK4 A0A026WAD8 A0A0L7QKE0 A0A088AJL8 A0A232EEI8 A0A154PQI5 A0A2P8YES0 U4UM22 A0A154PK09 A0A060GL20 A0A0M9AAI3 K7IQ63 J3JUU6 A0A170XK82 A0A0L7RCP2 A0A170XK51 E2ABY7 A0A1Y1LK10 A0A1W6EVW2 A0A1W7R8D5 A0A0C9S0C4 A0A0J7KGP1 A0A154P094 A0A154P3P1 A0A310SN00 A0A0C9RY44
EC Number
3.1.3.2
Pubmed
EMBL
NWSH01000024
PCG80631.1
ODYU01008500
SOQ52158.1
AGBW02010086
OWR49185.1
+ More
RSAL01000014 RVE53288.1 KQ460855 KPJ11938.1 KQ459601 KPI93943.1 BABH01004863 KPJ11936.1 KQ414714 KOC62926.1 KK853123 KDR11063.1 KPI93945.1 PCG80629.1 OWR49186.1 GDAI01002763 JAI14840.1 KQ971338 EFA02696.1 EFA02697.1 KQ971312 EEZ98635.1 ABLF02019532 DQ675545 ABG74711.1 KB632166 ERL89324.1 RVE53289.1 KY563421 ARK19830.1 APGK01027827 APGK01027828 BT128541 KB740671 KB631972 AEE63498.1 ENN79637.1 ERL87551.1 GL452008 EFN78070.1 GGMS01013238 MBY82441.1 GBHO01024763 JAG18841.1 GDHC01001938 JAQ16691.1 GL888120 EGI66977.1 LJIG01015968 KRT82010.1 QOIP01000003 RLU24852.1 ADTU01024956 ADTU01024957 KQ976719 KYM76739.1 GEZM01013380 GEZM01013379 JAV92604.1 GFDF01002423 JAV11661.1 GL442175 EFN63832.1 GL444789 EFN60592.1 GECZ01026189 JAS43580.1 EZ423150 ADD19426.1 GEBQ01005053 JAT34924.1 KZ288340 PBC27776.1 EFA02698.1 KQ762882 OAD55311.1 GFDF01002425 JAV11659.1 MF319183 AVZ66237.1 GFDL01015589 JAV19456.1 GFDL01015587 JAV19458.1 CVRI01000042 CRK95747.1 KQ981276 KYN43496.1 GL771866 EFZ09215.1 EU365869 ACA60733.1 GEZM01013669 JAV92386.1 GBYB01006952 JAG76719.1 GECU01004413 JAT03294.1 AY939855 FJ200211 DQ058012 GFDL01015588 JAV19457.1 EEZ98634.1 GFDL01015590 JAV19455.1 UFQS01001870 UFQT01001870 SSX12606.1 SSX32049.1 CCAG010002313 CCAG010002314 KZ288322 PBC28124.1 GAPW01001661 JAC11937.1 APGK01055292 APGK01055293 KB741261 ENN71644.1 EFN78071.1 JXJN01012024 KK107295 EZA53020.1 RLU24854.1 KQ414954 KOC59082.1 NNAY01005308 OXU16770.1 KQ435041 KZC14176.1 PYGN01000655 PSN42750.1 KB632278 ERL91221.1 KQ434943 KZC12199.1 KJ710422 AIB53032.1 KQ435699 KOX80575.1 BT127011 AEE61973.1 GEMB01004321 JAR98952.1 KQ414615 KOC68575.1 GEMB01004322 JAR98951.1 GL438386 EFN69082.1 GEZM01057530 JAV72275.1 KY563442 ARK19851.1 GEHC01000248 JAV47397.1 GBYB01001906 GBYB01015265 JAG71673.1 JAG85032.1 LBMM01007630 KMQ89563.1 KQ434787 KZC05263.1 KQ434796 KZC05750.1 KQ762196 OAD56135.1 GBYB01012831 JAG82598.1
RSAL01000014 RVE53288.1 KQ460855 KPJ11938.1 KQ459601 KPI93943.1 BABH01004863 KPJ11936.1 KQ414714 KOC62926.1 KK853123 KDR11063.1 KPI93945.1 PCG80629.1 OWR49186.1 GDAI01002763 JAI14840.1 KQ971338 EFA02696.1 EFA02697.1 KQ971312 EEZ98635.1 ABLF02019532 DQ675545 ABG74711.1 KB632166 ERL89324.1 RVE53289.1 KY563421 ARK19830.1 APGK01027827 APGK01027828 BT128541 KB740671 KB631972 AEE63498.1 ENN79637.1 ERL87551.1 GL452008 EFN78070.1 GGMS01013238 MBY82441.1 GBHO01024763 JAG18841.1 GDHC01001938 JAQ16691.1 GL888120 EGI66977.1 LJIG01015968 KRT82010.1 QOIP01000003 RLU24852.1 ADTU01024956 ADTU01024957 KQ976719 KYM76739.1 GEZM01013380 GEZM01013379 JAV92604.1 GFDF01002423 JAV11661.1 GL442175 EFN63832.1 GL444789 EFN60592.1 GECZ01026189 JAS43580.1 EZ423150 ADD19426.1 GEBQ01005053 JAT34924.1 KZ288340 PBC27776.1 EFA02698.1 KQ762882 OAD55311.1 GFDF01002425 JAV11659.1 MF319183 AVZ66237.1 GFDL01015589 JAV19456.1 GFDL01015587 JAV19458.1 CVRI01000042 CRK95747.1 KQ981276 KYN43496.1 GL771866 EFZ09215.1 EU365869 ACA60733.1 GEZM01013669 JAV92386.1 GBYB01006952 JAG76719.1 GECU01004413 JAT03294.1 AY939855 FJ200211 DQ058012 GFDL01015588 JAV19457.1 EEZ98634.1 GFDL01015590 JAV19455.1 UFQS01001870 UFQT01001870 SSX12606.1 SSX32049.1 CCAG010002313 CCAG010002314 KZ288322 PBC28124.1 GAPW01001661 JAC11937.1 APGK01055292 APGK01055293 KB741261 ENN71644.1 EFN78071.1 JXJN01012024 KK107295 EZA53020.1 RLU24854.1 KQ414954 KOC59082.1 NNAY01005308 OXU16770.1 KQ435041 KZC14176.1 PYGN01000655 PSN42750.1 KB632278 ERL91221.1 KQ434943 KZC12199.1 KJ710422 AIB53032.1 KQ435699 KOX80575.1 BT127011 AEE61973.1 GEMB01004321 JAR98952.1 KQ414615 KOC68575.1 GEMB01004322 JAR98951.1 GL438386 EFN69082.1 GEZM01057530 JAV72275.1 KY563442 ARK19851.1 GEHC01000248 JAV47397.1 GBYB01001906 GBYB01015265 JAG71673.1 JAG85032.1 LBMM01007630 KMQ89563.1 KQ434787 KZC05263.1 KQ434796 KZC05750.1 KQ762196 OAD56135.1 GBYB01012831 JAG82598.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
UP000005204
+ More
UP000053825 UP000027135 UP000002358 UP000007266 UP000007819 UP000030742 UP000079169 UP000019118 UP000008237 UP000007755 UP000279307 UP000005205 UP000078540 UP000000311 UP000242457 UP000183832 UP000078541 UP000092445 UP000091820 UP000005203 UP000092444 UP000092460 UP000053097 UP000215335 UP000076502 UP000245037 UP000053105 UP000036403
UP000053825 UP000027135 UP000002358 UP000007266 UP000007819 UP000030742 UP000079169 UP000019118 UP000008237 UP000007755 UP000279307 UP000005205 UP000078540 UP000000311 UP000242457 UP000183832 UP000078541 UP000092445 UP000091820 UP000005203 UP000092444 UP000092460 UP000053097 UP000215335 UP000076502 UP000245037 UP000053105 UP000036403
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K8T7
A0A2H1WGF6
A0A212F617
A0A3S2NZR1
A0A194R2C4
A0A194PLS5
+ More
H9J9I3 A0A194R7T8 A0A0L7QWI3 A0A067QNZ9 A0A194PS41 A0A2A4K986 A0A212F630 A0A0K8TLP0 K7IZV4 D2A1N0 D2A1N1 D6WAI2 J9K623 Q0PWU9 U4U626 A0A3S2LGG3 A0A1W6EVT0 A0A1S3D5K3 J3JZ60 A0A1S3D5S2 E2C1V8 A0A2S2QXE2 A0A0A9XGP6 A0A146MAA4 F4WFK0 A0A0T6B3P5 A0A3L8DX27 A0A158NSL2 A0A151HZE5 A0A1Y1N416 A0A1L8DYV9 K7IZV5 E2ARU9 E2B168 A0A1B6F055 D3TNZ8 A0A1B6MG81 A0A2A3E9C8 D2A1N2 A0A310SKW5 A0A1L8DZ06 A0A2R4SV17 A0A1Q3EW11 A0A1Q3EW78 A0A1J1I6C6 A0A195FSS6 E9JD78 C1IC99 A0A1A9ZSA2 A0A1Y1N7Z4 A0A1A9WGA5 K7IQ61 A0A0C9R287 A0A1B6JWK6 Q5BLY5 A0A1Q3EVW7 D6WAI1 A0A1Q3EVW8 A0A336L3V8 A0A087ZRC7 A0A1B0GD38 A0A2A3E9H3 A0A023EQR2 N6SVB5 E2C1V9 A0A1B0BCK4 A0A026WAD8 A0A0L7QKE0 A0A088AJL8 A0A232EEI8 A0A154PQI5 A0A2P8YES0 U4UM22 A0A154PK09 A0A060GL20 A0A0M9AAI3 K7IQ63 J3JUU6 A0A170XK82 A0A0L7RCP2 A0A170XK51 E2ABY7 A0A1Y1LK10 A0A1W6EVW2 A0A1W7R8D5 A0A0C9S0C4 A0A0J7KGP1 A0A154P094 A0A154P3P1 A0A310SN00 A0A0C9RY44
H9J9I3 A0A194R7T8 A0A0L7QWI3 A0A067QNZ9 A0A194PS41 A0A2A4K986 A0A212F630 A0A0K8TLP0 K7IZV4 D2A1N0 D2A1N1 D6WAI2 J9K623 Q0PWU9 U4U626 A0A3S2LGG3 A0A1W6EVT0 A0A1S3D5K3 J3JZ60 A0A1S3D5S2 E2C1V8 A0A2S2QXE2 A0A0A9XGP6 A0A146MAA4 F4WFK0 A0A0T6B3P5 A0A3L8DX27 A0A158NSL2 A0A151HZE5 A0A1Y1N416 A0A1L8DYV9 K7IZV5 E2ARU9 E2B168 A0A1B6F055 D3TNZ8 A0A1B6MG81 A0A2A3E9C8 D2A1N2 A0A310SKW5 A0A1L8DZ06 A0A2R4SV17 A0A1Q3EW11 A0A1Q3EW78 A0A1J1I6C6 A0A195FSS6 E9JD78 C1IC99 A0A1A9ZSA2 A0A1Y1N7Z4 A0A1A9WGA5 K7IQ61 A0A0C9R287 A0A1B6JWK6 Q5BLY5 A0A1Q3EVW7 D6WAI1 A0A1Q3EVW8 A0A336L3V8 A0A087ZRC7 A0A1B0GD38 A0A2A3E9H3 A0A023EQR2 N6SVB5 E2C1V9 A0A1B0BCK4 A0A026WAD8 A0A0L7QKE0 A0A088AJL8 A0A232EEI8 A0A154PQI5 A0A2P8YES0 U4UM22 A0A154PK09 A0A060GL20 A0A0M9AAI3 K7IQ63 J3JUU6 A0A170XK82 A0A0L7RCP2 A0A170XK51 E2ABY7 A0A1Y1LK10 A0A1W6EVW2 A0A1W7R8D5 A0A0C9S0C4 A0A0J7KGP1 A0A154P094 A0A154P3P1 A0A310SN00 A0A0C9RY44
PDB
1ND6
E-value=4.93219e-23,
Score=267
Ontologies
GO
PANTHER
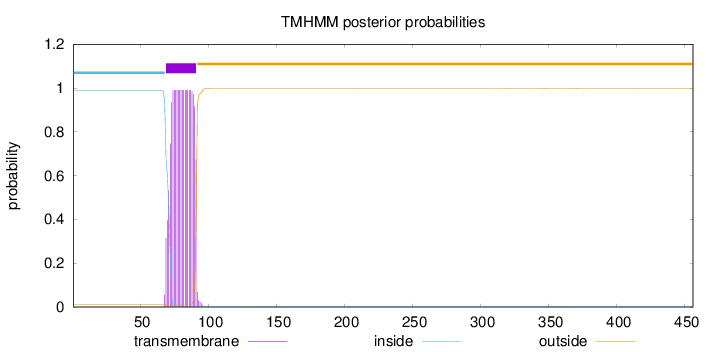
Topology
Subcellular location
Secreted
Length:
456
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.57503
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98871
inside
1 - 68
TMhelix
69 - 91
outside
92 - 456
Population Genetic Test Statistics
Pi
237.891324
Theta
168.421856
Tajima's D
0.904625
CLR
0.589076
CSRT
0.630418479076046
Interpretation
Uncertain
