Gene
KWMTBOMO01834 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006176
Annotation
PREDICTED:_venom_acid_phosphatase_Acph-1-like_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 3.267
Sequence
CDS
ATGAAACTATTCTCGACTGTATGGTTTCTGTTAGTCGTTAGTTGGTGCTCCGAAAGTACGCCCACGAGAAGGCTCGTCGAGCAGAATGTCGTGTCTGACAAGAGTCTACAGGATACCGATTTGCTCTTAACGTTCGTGGTATTTCGTCATGGCGACCGTACACCGGATCAAGAAGAACTGGCGTTGTTTCCGTCATCTACACAGGGCAATAAGGACTTGTTCTACCCTTACGGATTGAAGGCATTAACAAATATGGGTAAACGTCGAGCCTGTTCAGTCGGGAAATATCTCAGAAAGAACTATGATGGATTCCTTTCGAGGTTATACCTACCAGACGAGATTGTTATCCGCACTACGGATTATGATCGCACTAAAATGACAGCTTTAACTGCGATGGCCGGATTGTACCCACCAGAGCCGGAGCAACGATGGAACCCCACATTAAACTGGCAACCCGTCCCATATAACACTCCACCGCGCGATGAAGATGACTTACTATATTACTACAACTGCCCTCGTTATATTGCACTAAGAGATGAAACAGCTAGTTTGCCGGAAATCCAAGATTTATTGGCGCCGTACAAAGACCTCTTTCAATATTTAGAACAGCACACGAAAACTAATATTACTACATCGGAAGATGTTTTTTATTTAGACAATCTATTTCAAACACTGAGAAATGTCGGCGTTGAAACGCCAAAATGGGCACAGGAAGTGATGCCGCAAATAAAAGAAGTTACAAAAATCGAATATGCCATTCAGTATTATACTCCCGAATTGATAAGACTATCAGCTGGGGTTTTGTTAAACGAAATCTTAAATGCGACCGCTTCATATTTGTCCGGTGATACCGAGCAACATAAAGCACGTTTGTATTCGGCACACGAAAACAATGTAGCGGCAATAATGGCAGCCGCTAAAGTTTTCATACCTCATCAGCCTAATTACGGGTCTACGATAGCCATTGAATTCCGAAGGAATCGCACTACCGGAAAATATGGTTTCGCGGCGGTGTATGCATGCGATGCCGGCGGCCCGGGGAAGGTCCTCTCCGTAGACGGATGCGGCGGAGAAGCGTTTTGTGAATACGAAAAATTCGTCAATCTAGTCCAATCTTATAGAATTACATTAGAAGATTTTGAATACATCTGTCCGATATTAACTTAA
Protein
MKLFSTVWFLLVVSWCSESTPTRRLVEQNVVSDKSLQDTDLLLTFVVFRHGDRTPDQEELALFPSSTQGNKDLFYPYGLKALTNMGKRRACSVGKYLRKNYDGFLSRLYLPDEIVIRTTDYDRTKMTALTAMAGLYPPEPEQRWNPTLNWQPVPYNTPPRDEDDLLYYYNCPRYIALRDETASLPEIQDLLAPYKDLFQYLEQHTKTNITTSEDVFYLDNLFQTLRNVGVETPKWAQEVMPQIKEVTKIEYAIQYYTPELIRLSAGVLLNEILNATASYLSGDTEQHKARLYSAHENNVAAIMAAAKVFIPHQPNYGSTIAIEFRRNRTTGKYGFAAVYACDAGGPGKVLSVDGCGGEAFCEYEKFVNLVQSYRITLEDFEYICPILT
Summary
Similarity
Belongs to the histidine acid phosphatase family.
Uniprot
H9J9I3
A0A2A4K986
A0A3S2LGG3
A0A194R7T8
A0A194PS41
A0A2H1WGF6
+ More
A0A212F630 A0A0L7LP95 A0A1E1WMB7 S4NSI3 A0A212EPG3 A0A437AY25 A0A2A4JFS4 I4DR67 H9JIY7 A0A1E1W159 A0A1Y1N773 D2A1N1 A0A2A4K8T7 A0A0N0PBK2 A0A194PY67 U4U626 A0A3S2NZR1 E2C1V8 A0A195FSS6 K7IZV4 A0A026WAD8 A0A0J7KGP1 A0A158NSL2 F4WFK0 A0A151HZE5 A0A212F617 D2A1N2 J3JZC7 U4UM22 N6SZK1 C1IC99 U4UQB0 A0A194R2C4 E2B168 A0A195CQH0 J3JXC6 Q0PWU9 J3JZ60 A0A0M9AAI3 D2A1N0 A0A151WI99 E2B1D4 A0A1S3D5K3 A0A0L7QKE0 A0A0L7QWI3 A0A1S3D5S2 A0A154P094 A0A0C9S0C4 A0A1B6KL21 A0A154PK09 A0A0C9PLG4 K7IZV5 A0A1B6LZK6 A0A158P0J6 A0A2R4SV17 V5GYF9 A0A026WBJ4 D2A428 A0A0P5NSD8 A0A026WPS3 H9JXV1 A0A1Y1MDC4 A0A164T299 A0A0P5SWC6 A0A1Y1ME44 A0A0P5J485 A0A0P5QZ43 A0A151P351 A0A0P5H2M5 A0A0P5L5I5 A0A0P5PWV2 A0A0P5PNF8 A0A0P5NKF4 A0A0N8EIL9 A0A0P6D1E0 A0A0N8BMF4 A0A0N8DHR9 A0A0T6B1K6 E2B167 A0A310SN00 J3JY56 A0A0C9RY44 A0A2S2QXE2 A0A1L8DYV9 A0A1W6EVT0 A0A2H1W2H8 A0A0T6B4K7 K7IQ63 A0A1W4WJR1 B3NID9 B4PGG9 I4DJY6 A0A1U7RXA5
A0A212F630 A0A0L7LP95 A0A1E1WMB7 S4NSI3 A0A212EPG3 A0A437AY25 A0A2A4JFS4 I4DR67 H9JIY7 A0A1E1W159 A0A1Y1N773 D2A1N1 A0A2A4K8T7 A0A0N0PBK2 A0A194PY67 U4U626 A0A3S2NZR1 E2C1V8 A0A195FSS6 K7IZV4 A0A026WAD8 A0A0J7KGP1 A0A158NSL2 F4WFK0 A0A151HZE5 A0A212F617 D2A1N2 J3JZC7 U4UM22 N6SZK1 C1IC99 U4UQB0 A0A194R2C4 E2B168 A0A195CQH0 J3JXC6 Q0PWU9 J3JZ60 A0A0M9AAI3 D2A1N0 A0A151WI99 E2B1D4 A0A1S3D5K3 A0A0L7QKE0 A0A0L7QWI3 A0A1S3D5S2 A0A154P094 A0A0C9S0C4 A0A1B6KL21 A0A154PK09 A0A0C9PLG4 K7IZV5 A0A1B6LZK6 A0A158P0J6 A0A2R4SV17 V5GYF9 A0A026WBJ4 D2A428 A0A0P5NSD8 A0A026WPS3 H9JXV1 A0A1Y1MDC4 A0A164T299 A0A0P5SWC6 A0A1Y1ME44 A0A0P5J485 A0A0P5QZ43 A0A151P351 A0A0P5H2M5 A0A0P5L5I5 A0A0P5PWV2 A0A0P5PNF8 A0A0P5NKF4 A0A0N8EIL9 A0A0P6D1E0 A0A0N8BMF4 A0A0N8DHR9 A0A0T6B1K6 E2B167 A0A310SN00 J3JY56 A0A0C9RY44 A0A2S2QXE2 A0A1L8DYV9 A0A1W6EVT0 A0A2H1W2H8 A0A0T6B4K7 K7IQ63 A0A1W4WJR1 B3NID9 B4PGG9 I4DJY6 A0A1U7RXA5
Pubmed
EMBL
BABH01004863
NWSH01000024
PCG80629.1
RSAL01000014
RVE53289.1
KQ460855
+ More
KPJ11936.1 KQ459601 KPI93945.1 ODYU01008500 SOQ52158.1 AGBW02010086 OWR49186.1 JTDY01000422 KOB77267.1 GDQN01003058 JAT87996.1 GAIX01014082 JAA78478.1 AGBW02013469 OWR43369.1 RSAL01000278 RVE43078.1 NWSH01001685 PCG70424.1 AK404965 BAM20407.1 BABH01022999 BABH01023000 GDQN01010326 JAT80728.1 GEZM01011981 JAV93318.1 KQ971338 EFA02697.1 PCG80631.1 KQ460889 KPJ11041.1 KQ459585 KPI98277.1 KB632166 ERL89324.1 RVE53288.1 GL452008 EFN78070.1 KQ981276 KYN43496.1 KK107295 QOIP01000003 EZA53020.1 RLU24854.1 LBMM01007630 KMQ89563.1 ADTU01024956 ADTU01024957 GL888120 EGI66977.1 KQ976719 KYM76739.1 OWR49185.1 EFA02698.1 BT128608 AEE63565.1 KB632278 ERL91221.1 APGK01058345 KB741288 ENN70688.1 EU365869 ACA60733.1 KB632322 ERL92321.1 KPJ11938.1 GL444789 EFN60592.1 KQ977394 KYN02993.1 BT127895 AEE62857.1 ERL91264.1 DQ675545 ABG74711.1 APGK01027827 APGK01027828 BT128541 KB740671 KB631972 AEE63498.1 ENN79637.1 ERL87551.1 KQ435699 KOX80575.1 EFA02696.1 KQ983089 KYQ47583.1 GL444841 EFN60534.1 KQ414954 KOC59082.1 KQ414714 KOC62926.1 KQ434787 KZC05263.1 GBYB01001906 GBYB01015265 JAG71673.1 JAG85032.1 GEBQ01027828 JAT12149.1 KQ434943 KZC12199.1 GBYB01001903 JAG71670.1 GEBQ01010860 JAT29117.1 ADTU01005145 ADTU01005146 MF319183 AVZ66237.1 GALX01003038 JAB65428.1 EZA53021.1 RLU24853.1 KQ971348 EFA05605.1 GDIQ01142588 JAL09138.1 KK107139 EZA57636.1 RLU24559.1 BABH01040368 GEZM01036525 JAV82600.1 LRGB01001877 KZS10148.1 GDIP01139353 JAL64361.1 GEZM01036530 JAV82595.1 GDIQ01204904 JAK46821.1 GDIQ01108191 JAL43535.1 AKHW03001210 KYO43349.1 GDIQ01234056 JAK17669.1 GDIQ01183871 JAK67854.1 GDIQ01129285 JAL22441.1 GDIQ01146841 JAL04885.1 GDIQ01148236 JAL03490.1 GDIQ01023116 JAN71621.1 GDIQ01084352 JAN10385.1 GDIQ01163427 JAK88298.1 GDIP01032495 JAM71220.1 LJIG01016280 KRT81076.1 EFN60591.1 KQ762196 OAD56135.1 BT128180 AEE63141.1 GBYB01012831 JAG82598.1 GGMS01013238 MBY82441.1 GFDF01002423 JAV11661.1 KY563421 ARK19830.1 ODYU01005909 SOQ47289.1 LJIG01009939 KRT82078.1 CH954178 EDV52364.2 CM000159 EDW95328.2 AK401604 BAM18226.1
KPJ11936.1 KQ459601 KPI93945.1 ODYU01008500 SOQ52158.1 AGBW02010086 OWR49186.1 JTDY01000422 KOB77267.1 GDQN01003058 JAT87996.1 GAIX01014082 JAA78478.1 AGBW02013469 OWR43369.1 RSAL01000278 RVE43078.1 NWSH01001685 PCG70424.1 AK404965 BAM20407.1 BABH01022999 BABH01023000 GDQN01010326 JAT80728.1 GEZM01011981 JAV93318.1 KQ971338 EFA02697.1 PCG80631.1 KQ460889 KPJ11041.1 KQ459585 KPI98277.1 KB632166 ERL89324.1 RVE53288.1 GL452008 EFN78070.1 KQ981276 KYN43496.1 KK107295 QOIP01000003 EZA53020.1 RLU24854.1 LBMM01007630 KMQ89563.1 ADTU01024956 ADTU01024957 GL888120 EGI66977.1 KQ976719 KYM76739.1 OWR49185.1 EFA02698.1 BT128608 AEE63565.1 KB632278 ERL91221.1 APGK01058345 KB741288 ENN70688.1 EU365869 ACA60733.1 KB632322 ERL92321.1 KPJ11938.1 GL444789 EFN60592.1 KQ977394 KYN02993.1 BT127895 AEE62857.1 ERL91264.1 DQ675545 ABG74711.1 APGK01027827 APGK01027828 BT128541 KB740671 KB631972 AEE63498.1 ENN79637.1 ERL87551.1 KQ435699 KOX80575.1 EFA02696.1 KQ983089 KYQ47583.1 GL444841 EFN60534.1 KQ414954 KOC59082.1 KQ414714 KOC62926.1 KQ434787 KZC05263.1 GBYB01001906 GBYB01015265 JAG71673.1 JAG85032.1 GEBQ01027828 JAT12149.1 KQ434943 KZC12199.1 GBYB01001903 JAG71670.1 GEBQ01010860 JAT29117.1 ADTU01005145 ADTU01005146 MF319183 AVZ66237.1 GALX01003038 JAB65428.1 EZA53021.1 RLU24853.1 KQ971348 EFA05605.1 GDIQ01142588 JAL09138.1 KK107139 EZA57636.1 RLU24559.1 BABH01040368 GEZM01036525 JAV82600.1 LRGB01001877 KZS10148.1 GDIP01139353 JAL64361.1 GEZM01036530 JAV82595.1 GDIQ01204904 JAK46821.1 GDIQ01108191 JAL43535.1 AKHW03001210 KYO43349.1 GDIQ01234056 JAK17669.1 GDIQ01183871 JAK67854.1 GDIQ01129285 JAL22441.1 GDIQ01146841 JAL04885.1 GDIQ01148236 JAL03490.1 GDIQ01023116 JAN71621.1 GDIQ01084352 JAN10385.1 GDIQ01163427 JAK88298.1 GDIP01032495 JAM71220.1 LJIG01016280 KRT81076.1 EFN60591.1 KQ762196 OAD56135.1 BT128180 AEE63141.1 GBYB01012831 JAG82598.1 GGMS01013238 MBY82441.1 GFDF01002423 JAV11661.1 KY563421 ARK19830.1 ODYU01005909 SOQ47289.1 LJIG01009939 KRT82078.1 CH954178 EDV52364.2 CM000159 EDW95328.2 AK401604 BAM18226.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000007266 UP000030742 UP000008237 UP000078541 UP000002358 UP000053097 UP000279307 UP000036403 UP000005205 UP000007755 UP000078540 UP000019118 UP000000311 UP000078542 UP000053105 UP000075809 UP000079169 UP000053825 UP000076502 UP000076858 UP000050525 UP000192223 UP000008711 UP000002282 UP000189705
UP000037510 UP000007266 UP000030742 UP000008237 UP000078541 UP000002358 UP000053097 UP000279307 UP000036403 UP000005205 UP000007755 UP000078540 UP000019118 UP000000311 UP000078542 UP000053105 UP000075809 UP000079169 UP000053825 UP000076502 UP000076858 UP000050525 UP000192223 UP000008711 UP000002282 UP000189705
Interpro
Gene 3D
ProteinModelPortal
H9J9I3
A0A2A4K986
A0A3S2LGG3
A0A194R7T8
A0A194PS41
A0A2H1WGF6
+ More
A0A212F630 A0A0L7LP95 A0A1E1WMB7 S4NSI3 A0A212EPG3 A0A437AY25 A0A2A4JFS4 I4DR67 H9JIY7 A0A1E1W159 A0A1Y1N773 D2A1N1 A0A2A4K8T7 A0A0N0PBK2 A0A194PY67 U4U626 A0A3S2NZR1 E2C1V8 A0A195FSS6 K7IZV4 A0A026WAD8 A0A0J7KGP1 A0A158NSL2 F4WFK0 A0A151HZE5 A0A212F617 D2A1N2 J3JZC7 U4UM22 N6SZK1 C1IC99 U4UQB0 A0A194R2C4 E2B168 A0A195CQH0 J3JXC6 Q0PWU9 J3JZ60 A0A0M9AAI3 D2A1N0 A0A151WI99 E2B1D4 A0A1S3D5K3 A0A0L7QKE0 A0A0L7QWI3 A0A1S3D5S2 A0A154P094 A0A0C9S0C4 A0A1B6KL21 A0A154PK09 A0A0C9PLG4 K7IZV5 A0A1B6LZK6 A0A158P0J6 A0A2R4SV17 V5GYF9 A0A026WBJ4 D2A428 A0A0P5NSD8 A0A026WPS3 H9JXV1 A0A1Y1MDC4 A0A164T299 A0A0P5SWC6 A0A1Y1ME44 A0A0P5J485 A0A0P5QZ43 A0A151P351 A0A0P5H2M5 A0A0P5L5I5 A0A0P5PWV2 A0A0P5PNF8 A0A0P5NKF4 A0A0N8EIL9 A0A0P6D1E0 A0A0N8BMF4 A0A0N8DHR9 A0A0T6B1K6 E2B167 A0A310SN00 J3JY56 A0A0C9RY44 A0A2S2QXE2 A0A1L8DYV9 A0A1W6EVT0 A0A2H1W2H8 A0A0T6B4K7 K7IQ63 A0A1W4WJR1 B3NID9 B4PGG9 I4DJY6 A0A1U7RXA5
A0A212F630 A0A0L7LP95 A0A1E1WMB7 S4NSI3 A0A212EPG3 A0A437AY25 A0A2A4JFS4 I4DR67 H9JIY7 A0A1E1W159 A0A1Y1N773 D2A1N1 A0A2A4K8T7 A0A0N0PBK2 A0A194PY67 U4U626 A0A3S2NZR1 E2C1V8 A0A195FSS6 K7IZV4 A0A026WAD8 A0A0J7KGP1 A0A158NSL2 F4WFK0 A0A151HZE5 A0A212F617 D2A1N2 J3JZC7 U4UM22 N6SZK1 C1IC99 U4UQB0 A0A194R2C4 E2B168 A0A195CQH0 J3JXC6 Q0PWU9 J3JZ60 A0A0M9AAI3 D2A1N0 A0A151WI99 E2B1D4 A0A1S3D5K3 A0A0L7QKE0 A0A0L7QWI3 A0A1S3D5S2 A0A154P094 A0A0C9S0C4 A0A1B6KL21 A0A154PK09 A0A0C9PLG4 K7IZV5 A0A1B6LZK6 A0A158P0J6 A0A2R4SV17 V5GYF9 A0A026WBJ4 D2A428 A0A0P5NSD8 A0A026WPS3 H9JXV1 A0A1Y1MDC4 A0A164T299 A0A0P5SWC6 A0A1Y1ME44 A0A0P5J485 A0A0P5QZ43 A0A151P351 A0A0P5H2M5 A0A0P5L5I5 A0A0P5PWV2 A0A0P5PNF8 A0A0P5NKF4 A0A0N8EIL9 A0A0P6D1E0 A0A0N8BMF4 A0A0N8DHR9 A0A0T6B1K6 E2B167 A0A310SN00 J3JY56 A0A0C9RY44 A0A2S2QXE2 A0A1L8DYV9 A0A1W6EVT0 A0A2H1W2H8 A0A0T6B4K7 K7IQ63 A0A1W4WJR1 B3NID9 B4PGG9 I4DJY6 A0A1U7RXA5
PDB
2HPA
E-value=1.13316e-36,
Score=384
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.990482
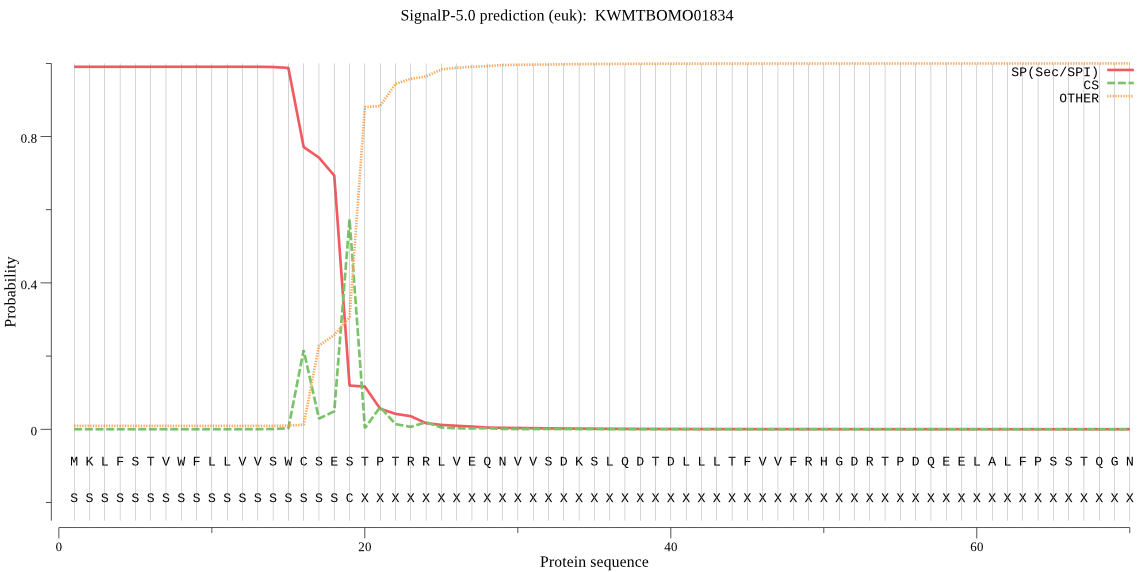
Length:
388
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01572
Exp number, first 60 AAs:
0.00375
Total prob of N-in:
0.00221
outside
1 - 388
Population Genetic Test Statistics
Pi
191.281706
Theta
193.313118
Tajima's D
0.185301
CLR
17.269932
CSRT
0.420878956052197
Interpretation
Uncertain
