Gene
KWMTBOMO01833
Pre Gene Modal
BGIBMGA005974
Annotation
PREDICTED:_dedicator_of_cytokinesis_protein_9_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.33
Sequence
CDS
ATGGCTGACGTGCATTACAACGAACAAGCATTGCTGGATCAGTTGTTGATATGCGCAGAATGTGCTGACAAGGCGGAGCGTTACGAACTCCTGGGTCCTTTGTATCGTCTCGTCATTCCGATCTACGAGAAAAGAAAGGACTATCAAGCGCTTTTAACTTGCTACCAGCATTTAACAAAAGCATACGCTAAAGTAATCGAAGTCACGAATTCAGGAAAAAGATTACTGGGCAGGTTTTACCGTGTCGCGTTCTTTGGGAAAGCGCATTTCGGTGAAGACGAAGAGGGTGTCGAATTCATATACAAGGAACCTAAACTGACGTCGCTCAGCGAAATTTCCGAGAACCTCCAAACGTTTTACGAACAAAAATTCGGTTGCGGCTTCGTTAAGATCATCATGGATTCGGCGCCCGTTAATCGCGAAGAACTAGACAGTAAGCTGGCCTACATCCAAGTTACATGGGTACGTGCGGCCCCAGAGGGGGTCACCACGACGGGCTCCAGCGCGGAGAGCTCGTTCGAACGCGTCCACAACGTGCGCAGATTCGTCTTCGAAACACCATTCACCAGCAGCGGCGCGGCCCGCGGACCAGTGCACGAACAGCACTTGCGCTTGACGCATCTAACAGCCGAAAACTGGTTTCCGTACGTGAAACGTCGCGTGCCCGTGATCGAGACGCAAGTTGAAGAGAAAAGTCCTATAGAGGTCGCTATCACCGAGATGGAGAGTCAAGTAATTGAACTCGCCGAAGTTGTCAACACGAAATCTCCAGATCTGAAGAAACTACAACTAAGATTGCAAGGAAGCGTGTGCGTACAAGTAAACGCCGGACCATTAGCGTACGCGAACGCATTCTTAGATCAAACTCTAGTCCACATGTACCCCGATGAGATGGTCGATAAATTGAAAGCCATATTTAAAGAATTTCTGACGGTGTGTCATTCAGCACTTCAACTCAACGCCAAACTGATAAACTCCGACCAGACGTCGTACCACGAAGCTCTCGAAAGTAATTACTACAAGCTCAGCGACTCCCTGTCGTCCGTGCTCGGGCAGCCGCTGCACGATATACTCGTCAACGGGACCCTCAGTACCACGGTCACCGGTGTAGAAATCGAATCTAGTAACGCTTAA
Protein
MADVHYNEQALLDQLLICAECADKAERYELLGPLYRLVIPIYEKRKDYQALLTCYQHLTKAYAKVIEVTNSGKRLLGRFYRVAFFGKAHFGEDEEGVEFIYKEPKLTSLSEISENLQTFYEQKFGCGFVKIIMDSAPVNREELDSKLAYIQVTWVRAAPEGVTTTGSSAESSFERVHNVRRFVFETPFTSSGAARGPVHEQHLRLTHLTAENWFPYVKRRVPVIETQVEEKSPIEVAITEMESQVIELAEVVNTKSPDLKKLQLRLQGSVCVQVNAGPLAYANAFLDQTLVHMYPDEMVDKLKAIFKEFLTVCHSALQLNAKLINSDQTSYHEALESNYYKLSDSLSSVLGQPLHDILVNGTLSTTVTGVEIESSNA
Summary
Similarity
Belongs to the DOCK family.
Uniprot
H9J8Y2
A0A212F624
A0A2A4K9Q4
A0A2A4K8U3
A0A194R3W7
A0A437BSD0
+ More
A0A194PKI2 A0A2H1WGK0 A0A2J7R7J8 A0A2R7WDQ1 A0A310SEX8 A0A0J7N863 A0A088AQN5 A0A154NX43 E9IW93 A0A2A3EH22 F4X2S7 A0A151IAP8 A0A195BLI2 E2A4X4 A0A158NLQ7 E2BSG6 A0A151IU48 A0A195FVS9 A0A151WK12 A0A026WF93 A0A3L8DIN0 A0A2J7R7J9 A0A067RL42 A0A1B6EDB7 A0A0C9Q184 A0A1B6I6N9 A0A0C9R4D2 A0A1B6I8Z2 A0A0C9RH84 A0A232FFG2 A0A146LQ50 A0A146LXB0 A0A1W4X9S1 U4U1X9 N6TER4 W5JRI3 W4VS04 A0A139WGV9 D6WLQ6 A0A0T6B0Z6 A0A0A1XB92 A0A182JHE5 A0A182FNB8 M7BNI8 A0A182VSH9 A0A2H6MXK4 A0A0L0CGE9 A0A2H6MXI8 A0A182YP55 A0A0S7JL51 A0A2D4KRZ5 C3ZF33 A0A0K8UB62 Q7Z6H4 A0A182MBY6 A0A0S7JMM5 B0BM88 A0A182RWD5 A0A034V6U3 A0A0S7JL58 A0A0K8TZR3 A0A182Q331 A0A1B0D1K7 A0A0S7JN80 A0A2D4Q520 A0A0P6ITV5 A0A034V3H9 A0A0A1WGY8 A0A0L7RKD5 W8B9Y8 W8AEL3 W8B0C6 A0A182NT68 Q6PF84 A0A023F2T5 G3UTR5 A0A1I8PQ27 A0A3B4E013 A0A1I8PPX3 A0A1I8PQ35 A0A182K3W5 A0A1I8ME53 A0A1I8PPX8 A0A1I8PPX0 A0A1I8PQ16 A0A1I8PPW5 A0A1I8PQ33 T1H8L3 H2YGG2 A0A2K6FNT2 A0A1I8PQ24 M1ELN5 A0A182P4N5 B7Z6G9
A0A194PKI2 A0A2H1WGK0 A0A2J7R7J8 A0A2R7WDQ1 A0A310SEX8 A0A0J7N863 A0A088AQN5 A0A154NX43 E9IW93 A0A2A3EH22 F4X2S7 A0A151IAP8 A0A195BLI2 E2A4X4 A0A158NLQ7 E2BSG6 A0A151IU48 A0A195FVS9 A0A151WK12 A0A026WF93 A0A3L8DIN0 A0A2J7R7J9 A0A067RL42 A0A1B6EDB7 A0A0C9Q184 A0A1B6I6N9 A0A0C9R4D2 A0A1B6I8Z2 A0A0C9RH84 A0A232FFG2 A0A146LQ50 A0A146LXB0 A0A1W4X9S1 U4U1X9 N6TER4 W5JRI3 W4VS04 A0A139WGV9 D6WLQ6 A0A0T6B0Z6 A0A0A1XB92 A0A182JHE5 A0A182FNB8 M7BNI8 A0A182VSH9 A0A2H6MXK4 A0A0L0CGE9 A0A2H6MXI8 A0A182YP55 A0A0S7JL51 A0A2D4KRZ5 C3ZF33 A0A0K8UB62 Q7Z6H4 A0A182MBY6 A0A0S7JMM5 B0BM88 A0A182RWD5 A0A034V6U3 A0A0S7JL58 A0A0K8TZR3 A0A182Q331 A0A1B0D1K7 A0A0S7JN80 A0A2D4Q520 A0A0P6ITV5 A0A034V3H9 A0A0A1WGY8 A0A0L7RKD5 W8B9Y8 W8AEL3 W8B0C6 A0A182NT68 Q6PF84 A0A023F2T5 G3UTR5 A0A1I8PQ27 A0A3B4E013 A0A1I8PPX3 A0A1I8PQ35 A0A182K3W5 A0A1I8ME53 A0A1I8PPX8 A0A1I8PPX0 A0A1I8PQ16 A0A1I8PPW5 A0A1I8PQ33 T1H8L3 H2YGG2 A0A2K6FNT2 A0A1I8PQ24 M1ELN5 A0A182P4N5 B7Z6G9
Pubmed
EMBL
BABH01004863
AGBW02010086
OWR49187.1
NWSH01000024
PCG80628.1
PCG80627.1
+ More
KQ460855 KPJ11935.1 RSAL01000014 RVE53290.1 KQ459601 KPI93946.1 ODYU01008500 SOQ52157.1 NEVH01006731 PNF36809.1 KK854512 PTY16475.1 KQ769580 OAD52855.1 LBMM01008499 KMQ88825.1 KQ434777 KZC04246.1 GL766458 EFZ15167.1 KZ288253 PBC31020.1 GL888591 EGI59273.1 KQ978186 KYM96573.1 KQ976450 KYM85662.1 GL436756 EFN71516.1 ADTU01019667 GL450233 EFN81354.1 KQ980968 KYN10954.1 KQ981219 KYN44412.1 KQ983020 KYQ48202.1 KK107238 EZA54722.1 QOIP01000007 RLU20211.1 PNF36810.1 KK852561 KDR21325.1 GEDC01001377 JAS35921.1 GBYB01007668 JAG77435.1 GECU01025115 JAS82591.1 GBYB01007667 JAG77434.1 GECU01024301 JAS83405.1 GBYB01007665 JAG77432.1 NNAY01000330 OXU29169.1 GDHC01010119 JAQ08510.1 GDHC01006361 GDHC01003287 JAQ12268.1 JAQ15342.1 KB631904 ERL87077.1 APGK01040726 APGK01040727 KB740984 ENN76248.1 ADMH02000390 ETN66746.1 GANO01000103 JAB59768.1 KQ971343 KYB27228.1 EFA03420.2 LJIG01016254 KRT81152.1 GBXI01005658 JAD08634.1 KB554236 EMP29752.1 IACI01013235 LAA19898.1 JRES01000438 KNC31320.1 IACI01013237 LAA19902.1 GBYX01236038 JAO65937.1 IACL01080845 LAB11485.1 GG666612 EEN49339.1 GDHF01028508 GDHF01013159 JAI23806.1 JAI39155.1 BC053620 AAH53620.1 AXCM01002695 GBYX01236037 JAO65938.1 BC158330 AAI58331.1 GAKP01021110 JAC37842.1 GBYX01236033 JAO65942.1 GDHF01032568 JAI19746.1 AXCN02000728 AJVK01010287 AJVK01010288 AJVK01010289 AJVK01010290 GBYX01236027 JAO65947.1 IACN01111071 LAB65332.1 GDUN01000676 JAN95243.1 GAKP01021111 JAC37841.1 GBXI01016023 JAC98268.1 KQ414578 KOC71206.1 GAMC01019911 JAB86644.1 GAMC01019910 JAB86645.1 GAMC01019909 JAB86646.1 BC057692 AAH57692.1 GBBI01003209 JAC15503.1 ACPB03001107 ACPB03001108 ACPB03001109 JP008882 AER97479.1 AK300298 BAH13255.1
KQ460855 KPJ11935.1 RSAL01000014 RVE53290.1 KQ459601 KPI93946.1 ODYU01008500 SOQ52157.1 NEVH01006731 PNF36809.1 KK854512 PTY16475.1 KQ769580 OAD52855.1 LBMM01008499 KMQ88825.1 KQ434777 KZC04246.1 GL766458 EFZ15167.1 KZ288253 PBC31020.1 GL888591 EGI59273.1 KQ978186 KYM96573.1 KQ976450 KYM85662.1 GL436756 EFN71516.1 ADTU01019667 GL450233 EFN81354.1 KQ980968 KYN10954.1 KQ981219 KYN44412.1 KQ983020 KYQ48202.1 KK107238 EZA54722.1 QOIP01000007 RLU20211.1 PNF36810.1 KK852561 KDR21325.1 GEDC01001377 JAS35921.1 GBYB01007668 JAG77435.1 GECU01025115 JAS82591.1 GBYB01007667 JAG77434.1 GECU01024301 JAS83405.1 GBYB01007665 JAG77432.1 NNAY01000330 OXU29169.1 GDHC01010119 JAQ08510.1 GDHC01006361 GDHC01003287 JAQ12268.1 JAQ15342.1 KB631904 ERL87077.1 APGK01040726 APGK01040727 KB740984 ENN76248.1 ADMH02000390 ETN66746.1 GANO01000103 JAB59768.1 KQ971343 KYB27228.1 EFA03420.2 LJIG01016254 KRT81152.1 GBXI01005658 JAD08634.1 KB554236 EMP29752.1 IACI01013235 LAA19898.1 JRES01000438 KNC31320.1 IACI01013237 LAA19902.1 GBYX01236038 JAO65937.1 IACL01080845 LAB11485.1 GG666612 EEN49339.1 GDHF01028508 GDHF01013159 JAI23806.1 JAI39155.1 BC053620 AAH53620.1 AXCM01002695 GBYX01236037 JAO65938.1 BC158330 AAI58331.1 GAKP01021110 JAC37842.1 GBYX01236033 JAO65942.1 GDHF01032568 JAI19746.1 AXCN02000728 AJVK01010287 AJVK01010288 AJVK01010289 AJVK01010290 GBYX01236027 JAO65947.1 IACN01111071 LAB65332.1 GDUN01000676 JAN95243.1 GAKP01021111 JAC37841.1 GBXI01016023 JAC98268.1 KQ414578 KOC71206.1 GAMC01019911 JAB86644.1 GAMC01019910 JAB86645.1 GAMC01019909 JAB86646.1 BC057692 AAH57692.1 GBBI01003209 JAC15503.1 ACPB03001107 ACPB03001108 ACPB03001109 JP008882 AER97479.1 AK300298 BAH13255.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000283053
UP000053268
+ More
UP000235965 UP000036403 UP000005203 UP000076502 UP000242457 UP000007755 UP000078542 UP000078540 UP000000311 UP000005205 UP000008237 UP000078492 UP000078541 UP000075809 UP000053097 UP000279307 UP000027135 UP000215335 UP000192223 UP000030742 UP000019118 UP000000673 UP000007266 UP000075880 UP000069272 UP000031443 UP000075920 UP000037069 UP000076408 UP000001554 UP000075883 UP000075900 UP000075886 UP000092462 UP000053825 UP000075884 UP000001645 UP000095300 UP000261440 UP000075881 UP000095301 UP000015103 UP000007875 UP000233160 UP000075885
UP000235965 UP000036403 UP000005203 UP000076502 UP000242457 UP000007755 UP000078542 UP000078540 UP000000311 UP000005205 UP000008237 UP000078492 UP000078541 UP000075809 UP000053097 UP000279307 UP000027135 UP000215335 UP000192223 UP000030742 UP000019118 UP000000673 UP000007266 UP000075880 UP000069272 UP000031443 UP000075920 UP000037069 UP000076408 UP000001554 UP000075883 UP000075900 UP000075886 UP000092462 UP000053825 UP000075884 UP000001645 UP000095300 UP000261440 UP000075881 UP000095301 UP000015103 UP000007875 UP000233160 UP000075885
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9J8Y2
A0A212F624
A0A2A4K9Q4
A0A2A4K8U3
A0A194R3W7
A0A437BSD0
+ More
A0A194PKI2 A0A2H1WGK0 A0A2J7R7J8 A0A2R7WDQ1 A0A310SEX8 A0A0J7N863 A0A088AQN5 A0A154NX43 E9IW93 A0A2A3EH22 F4X2S7 A0A151IAP8 A0A195BLI2 E2A4X4 A0A158NLQ7 E2BSG6 A0A151IU48 A0A195FVS9 A0A151WK12 A0A026WF93 A0A3L8DIN0 A0A2J7R7J9 A0A067RL42 A0A1B6EDB7 A0A0C9Q184 A0A1B6I6N9 A0A0C9R4D2 A0A1B6I8Z2 A0A0C9RH84 A0A232FFG2 A0A146LQ50 A0A146LXB0 A0A1W4X9S1 U4U1X9 N6TER4 W5JRI3 W4VS04 A0A139WGV9 D6WLQ6 A0A0T6B0Z6 A0A0A1XB92 A0A182JHE5 A0A182FNB8 M7BNI8 A0A182VSH9 A0A2H6MXK4 A0A0L0CGE9 A0A2H6MXI8 A0A182YP55 A0A0S7JL51 A0A2D4KRZ5 C3ZF33 A0A0K8UB62 Q7Z6H4 A0A182MBY6 A0A0S7JMM5 B0BM88 A0A182RWD5 A0A034V6U3 A0A0S7JL58 A0A0K8TZR3 A0A182Q331 A0A1B0D1K7 A0A0S7JN80 A0A2D4Q520 A0A0P6ITV5 A0A034V3H9 A0A0A1WGY8 A0A0L7RKD5 W8B9Y8 W8AEL3 W8B0C6 A0A182NT68 Q6PF84 A0A023F2T5 G3UTR5 A0A1I8PQ27 A0A3B4E013 A0A1I8PPX3 A0A1I8PQ35 A0A182K3W5 A0A1I8ME53 A0A1I8PPX8 A0A1I8PPX0 A0A1I8PQ16 A0A1I8PPW5 A0A1I8PQ33 T1H8L3 H2YGG2 A0A2K6FNT2 A0A1I8PQ24 M1ELN5 A0A182P4N5 B7Z6G9
A0A194PKI2 A0A2H1WGK0 A0A2J7R7J8 A0A2R7WDQ1 A0A310SEX8 A0A0J7N863 A0A088AQN5 A0A154NX43 E9IW93 A0A2A3EH22 F4X2S7 A0A151IAP8 A0A195BLI2 E2A4X4 A0A158NLQ7 E2BSG6 A0A151IU48 A0A195FVS9 A0A151WK12 A0A026WF93 A0A3L8DIN0 A0A2J7R7J9 A0A067RL42 A0A1B6EDB7 A0A0C9Q184 A0A1B6I6N9 A0A0C9R4D2 A0A1B6I8Z2 A0A0C9RH84 A0A232FFG2 A0A146LQ50 A0A146LXB0 A0A1W4X9S1 U4U1X9 N6TER4 W5JRI3 W4VS04 A0A139WGV9 D6WLQ6 A0A0T6B0Z6 A0A0A1XB92 A0A182JHE5 A0A182FNB8 M7BNI8 A0A182VSH9 A0A2H6MXK4 A0A0L0CGE9 A0A2H6MXI8 A0A182YP55 A0A0S7JL51 A0A2D4KRZ5 C3ZF33 A0A0K8UB62 Q7Z6H4 A0A182MBY6 A0A0S7JMM5 B0BM88 A0A182RWD5 A0A034V6U3 A0A0S7JL58 A0A0K8TZR3 A0A182Q331 A0A1B0D1K7 A0A0S7JN80 A0A2D4Q520 A0A0P6ITV5 A0A034V3H9 A0A0A1WGY8 A0A0L7RKD5 W8B9Y8 W8AEL3 W8B0C6 A0A182NT68 Q6PF84 A0A023F2T5 G3UTR5 A0A1I8PQ27 A0A3B4E013 A0A1I8PPX3 A0A1I8PQ35 A0A182K3W5 A0A1I8ME53 A0A1I8PPX8 A0A1I8PPX0 A0A1I8PQ16 A0A1I8PPW5 A0A1I8PQ33 T1H8L3 H2YGG2 A0A2K6FNT2 A0A1I8PQ24 M1ELN5 A0A182P4N5 B7Z6G9
PDB
2WMO
E-value=5.49147e-94,
Score=878
Ontologies
PANTHER
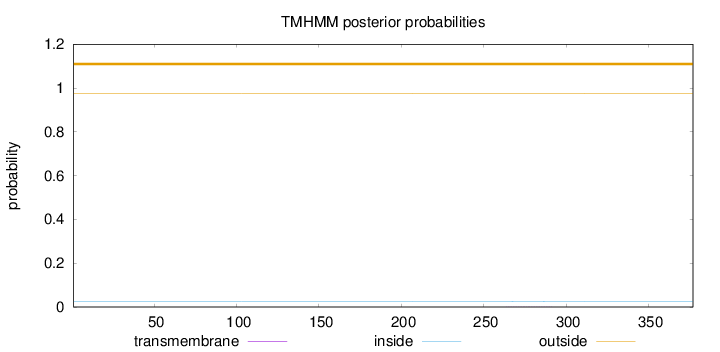
Topology
Length:
377
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01139
Exp number, first 60 AAs:
0.00049
Total prob of N-in:
0.02471
outside
1 - 377
Population Genetic Test Statistics
Pi
194.376069
Theta
143.728503
Tajima's D
1.007429
CLR
0
CSRT
0.662116894155292
Interpretation
Uncertain
