Gene
KWMTBOMO01832
Pre Gene Modal
BGIBMGA005975
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_phosphatase_1L_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.24
Sequence
CDS
ATGGAAGACGAGATAGAAGACAGAGTATTACATCAAACGTACGTTTCTTTTGTAAAAATATTGTCTAGATTCACCAGTAGCACTTTTCTTGACGCTCCTGTGAGCTATTTATGGAAAATGGTGCGTCTTTATTTTCTGCGATCTGAAGTAGTTGTGATCACACTAGCTATTGTGATCTTCTTAATGTATCTTCAAAGTATAGAATTATGGAGTCGAACAATGCTAAGCAGACTGTCGCAAGCGATGAATCCTATGAGCGCTGTTCAACGAATGAAACTCATGGAGGGATCCGATGCAGACAAGCAAAGTTGGGAACTGAAAGGCACGTTAAGTGCTGCTTACGCAATCAAAGGTAGAAGAATGCACATGGAAGATAGATTTATAATAAACGAAAATGTAAACAACACAGGCATATCACTATTCGCTATATTCGATGGACACGGCGGAGAGTTTGCTGCTAACTATGCCAAGGACCATCTTATACAGAATCTTTACAACAAAATTGTTGAACTTAACGCTTTCAAAGAAGGTAAAATATCTGATATATCTAGAGAAATTATTGAGGAGTATGCACCTAAAGAGGAAAATGTTGAACGTAAAACAAGCTTCAAAAAGTCAGTTAGTACTGCAGATGATACATCAAAAAAAGAAATAACAGATCCTGAATTGCTTGCCCAGCTTTCTAAAGCAAGGCCAATCATTACCCGAGAAGTGCGGACCACAACTAAGCCAGTTAAGAATATCAATATACCATTATCTAGTTATATTGACAAAGGAAAAATTAATTATGGAAAATTATTGACTGATGAAGTTTTAGCAGCAGATAGATTACTAGTAGAAGCTGCAAAGAAGTCTTTGAATGTTGCCGGCACTACAGCCCTTATTGCAATAATGGAAAATAATCATCTGATAGTGGCCAATGTTGGTGACTCCAGGGGAGTCATGTGTGACTCGAGAGGCAATGCTATTCCTGTCTCGTTTGACCATAAACCACAACAGGTAAGGGAACAAAAAAGAATTGAGGCAGCAGGAGGTTACATTGCTTATAATGGAGTTTGGAGAGTTGCAGGTATCTTAGCTACTTCAAGAGCTATGGGTGATTACCCACTCAAAGATAAGAAATTTGTCATAGCAGATCCAGATATACTTACTTTTAACTTAAATGATCATAAACCTATGTTCTTAGTCTTAGCCTCTGATGGTCTCTGGGACACGTTTACCAATGAAGAAGCTGTGAAATTTATTAAAGAAAGACTTGATGAACCAGATTATGGGGCAAAAAGTTTAACACTACAAGCATATTATAGAGGATCAGTCGATAATATTACAGTTTTGGTCATAAACTTCCTAAATACCAATATAACCATCGCAGGAGTTCAGTAG
Protein
MEDEIEDRVLHQTYVSFVKILSRFTSSTFLDAPVSYLWKMVRLYFLRSEVVVITLAIVIFLMYLQSIELWSRTMLSRLSQAMNPMSAVQRMKLMEGSDADKQSWELKGTLSAAYAIKGRRMHMEDRFIINENVNNTGISLFAIFDGHGGEFAANYAKDHLIQNLYNKIVELNAFKEGKISDISREIIEEYAPKEENVERKTSFKKSVSTADDTSKKEITDPELLAQLSKARPIITREVRTTTKPVKNINIPLSSYIDKGKINYGKLLTDEVLAADRLLVEAAKKSLNVAGTTALIAIMENNHLIVANVGDSRGVMCDSRGNAIPVSFDHKPQQVREQKRIEAAGGYIAYNGVWRVAGILATSRAMGDYPLKDKKFVIADPDILTFNLNDHKPMFLVLASDGLWDTFTNEEAVKFIKERLDEPDYGAKSLTLQAYYRGSVDNITVLVINFLNTNITIAGVQ
Summary
Cofactor
Mg(2+)
Similarity
Belongs to the PP2C family.
Uniprot
A0A3S2NPY3
A0A2H1WGS4
A0A2A4K9H8
A0A1E1W2Z7
A0A194R337
A0A194PKU1
+ More
A0A212F632 A0A0L7LNZ4 H9J8Y3 A0A2J7R7K9 D6WLQ8 A0A2P8Y7P6 A0A1Y1L5Q3 A0A067RBQ9 A0A1B6ILH2 A0A1B6M308 A0A1W4X9I7 A0A1B6GD57 A0A088AQK8 A0A2A3EII8 V9IFR0 A0A3L8DDZ1 A0A154NX95 A0A0T6B1A8 A0A0L7RK12 A0A232FH79 A0A0C9R2K5 E2AKW4 A0A1B6CPH1 A0A026X3F9 A0A0P4VTL3 R4G3X5 A0A069DTV7 A0A0V0G3D6 A0A224XPJ5 A0A023F8K4 A0A0A9WXK6 R4WRP8 A0A0K8TR79 A0A0L0CIL8 A0A310S5L4 A0A161M4M2 A0A2R7WA99 A0A1B0AIU1 A0A3Q0J0B5 A0A1A9X8A1 A0A1B0C441 A0A1I8N0K0 T1PFI6 A0A1I8P0Q7 A0A0M3QTI3 U4U415 B3MV14 N6T7P8 B4JEI1 A0A1W4W8I8 A0A1A9WBW6 B4KG60 W8B689 A0A0K8VVC3 A0A034W9I4 B4LSS0 E0VF87 A0A1L8DJT2 A0A182MMC8 A0A0A1XPR0 A0A3B0JES6 B4MWK3 B4G9I2 Q29MA9 U5EEM8 A0A182VSH7 A0A182L024 B4NZG2 A0A0R1DJE4 A0A1B0CLI3 A0A2M4BKG9 A0A182FNB6 Q9XZ28 Q17C45 A0A182K1K7 A0A2M4AD20 B3N6P4 A0A182RWD2 A0A0J9QY22 B4HYB0 A0A2M4ADB2 W5JTB7 A0A2M3Z6V6 A0A1Q3FH28 A0A1Q3FH01 A0A182TMZ4 B4Q5U1 A0A182NT70 A0A182IBS9 Q7Q4Q8 A0A182QRH9 A0A182X198 A0A182UV48 A0A182YP54
A0A212F632 A0A0L7LNZ4 H9J8Y3 A0A2J7R7K9 D6WLQ8 A0A2P8Y7P6 A0A1Y1L5Q3 A0A067RBQ9 A0A1B6ILH2 A0A1B6M308 A0A1W4X9I7 A0A1B6GD57 A0A088AQK8 A0A2A3EII8 V9IFR0 A0A3L8DDZ1 A0A154NX95 A0A0T6B1A8 A0A0L7RK12 A0A232FH79 A0A0C9R2K5 E2AKW4 A0A1B6CPH1 A0A026X3F9 A0A0P4VTL3 R4G3X5 A0A069DTV7 A0A0V0G3D6 A0A224XPJ5 A0A023F8K4 A0A0A9WXK6 R4WRP8 A0A0K8TR79 A0A0L0CIL8 A0A310S5L4 A0A161M4M2 A0A2R7WA99 A0A1B0AIU1 A0A3Q0J0B5 A0A1A9X8A1 A0A1B0C441 A0A1I8N0K0 T1PFI6 A0A1I8P0Q7 A0A0M3QTI3 U4U415 B3MV14 N6T7P8 B4JEI1 A0A1W4W8I8 A0A1A9WBW6 B4KG60 W8B689 A0A0K8VVC3 A0A034W9I4 B4LSS0 E0VF87 A0A1L8DJT2 A0A182MMC8 A0A0A1XPR0 A0A3B0JES6 B4MWK3 B4G9I2 Q29MA9 U5EEM8 A0A182VSH7 A0A182L024 B4NZG2 A0A0R1DJE4 A0A1B0CLI3 A0A2M4BKG9 A0A182FNB6 Q9XZ28 Q17C45 A0A182K1K7 A0A2M4AD20 B3N6P4 A0A182RWD2 A0A0J9QY22 B4HYB0 A0A2M4ADB2 W5JTB7 A0A2M3Z6V6 A0A1Q3FH28 A0A1Q3FH01 A0A182TMZ4 B4Q5U1 A0A182NT70 A0A182IBS9 Q7Q4Q8 A0A182QRH9 A0A182X198 A0A182UV48 A0A182YP54
Pubmed
26354079
22118469
26227816
19121390
18362917
19820115
+ More
29403074 28004739 24845553 30249741 28648823 20798317 24508170 27129103 26334808 25474469 25401762 26823975 23691247 26369729 26108605 25315136 23537049 17994087 18057021 24495485 25348373 20566863 25830018 15632085 20966253 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 22936249 20920257 23761445 12364791 14747013 17210077 25244985
29403074 28004739 24845553 30249741 28648823 20798317 24508170 27129103 26334808 25474469 25401762 26823975 23691247 26369729 26108605 25315136 23537049 17994087 18057021 24495485 25348373 20566863 25830018 15632085 20966253 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 22936249 20920257 23761445 12364791 14747013 17210077 25244985
EMBL
RSAL01000014
RVE53291.1
ODYU01008500
SOQ52156.1
NWSH01000024
PCG80626.1
+ More
GDQN01009701 JAT81353.1 KQ460855 KPJ11934.1 KQ459601 KPI93947.1 AGBW02010086 OWR49188.1 JTDY01000422 KOB77258.1 BABH01004861 BABH01004862 NEVH01006731 PNF36811.1 KQ971343 EFA04151.1 PYGN01000830 PSN40265.1 GEZM01063934 JAV68904.1 KK852561 KDR21326.1 GECU01019935 JAS87771.1 GEBQ01009656 JAT30321.1 GECZ01009405 JAS60364.1 KZ288253 PBC31019.1 JR044938 AEY59920.1 QOIP01000009 RLU18516.1 KQ434777 KZC04247.1 LJIG01016254 KRT81153.1 KQ414578 KOC71205.1 NNAY01000193 OXU30116.1 GBYB01007072 JAG76839.1 GL440425 EFN65881.1 GEDC01021970 JAS15328.1 KK107038 EZA61934.1 GDKW01001278 JAI55317.1 GAHY01001286 JAA76224.1 GBGD01001361 JAC87528.1 GECL01003519 JAP02605.1 GFTR01006507 JAW09919.1 GBBI01000996 JAC17716.1 GBHO01035408 GBHO01035407 GBHO01030436 GBHO01030422 GBRD01002069 GBRD01002068 GDHC01015666 GDHC01012446 GDHC01003077 JAG08196.1 JAG08197.1 JAG13168.1 JAG13182.1 JAG63752.1 JAQ02963.1 JAQ06183.1 JAQ15552.1 AK417372 BAN20587.1 GDAI01000719 JAI16884.1 JRES01000438 KNC31324.1 KQ769580 OAD52854.1 GEMB01002826 JAS00373.1 KK854512 PTY16469.1 JXJN01025268 KA647557 AFP62186.1 CP012523 ALC38939.1 KB631904 ERL87078.1 CH902624 EDV33079.1 KPU74290.1 APGK01040727 KB740984 ENN76249.1 CH916368 EDW03701.1 CH933807 EDW13199.1 GAMC01012441 GAMC01012440 GAMC01012439 GAMC01012438 JAB94117.1 GDHF01009683 GDHF01009579 GDHF01009495 JAI42631.1 JAI42735.1 JAI42819.1 GAKP01007973 GAKP01007972 GAKP01007970 GAKP01007968 JAC50982.1 CH940649 EDW63809.1 DS235107 EEB12043.1 GFDF01007345 JAV06739.1 AXCM01001483 GBXI01015359 GBXI01001457 JAC98932.1 JAD12835.1 OUUW01000004 SPP79163.1 CH963857 EDW76144.1 CH479180 EDW29012.1 CH379060 EAL33785.1 GANO01004251 JAB55620.1 CM000157 EDW87711.1 KRJ97323.1 KRJ97322.1 AJWK01017307 GGFJ01004419 MBW53560.1 AF132185 AE014134 AAD34773.1 AAF52564.1 AAF52565.1 AGB92752.1 CH477313 EAT43857.1 GGFK01005364 MBW38685.1 CH954177 EDV59260.1 CM002910 KMY88863.1 CH480818 EDW52040.1 GGFK01005442 MBW38763.1 ADMH02000497 ETN66195.1 GGFM01003469 MBW24220.1 GFDL01008188 JAV26857.1 GFDL01008195 JAV26850.1 CM000361 EDX04137.1 APCN01000736 AAAB01008964 EAA12153.4 AXCN02000728
GDQN01009701 JAT81353.1 KQ460855 KPJ11934.1 KQ459601 KPI93947.1 AGBW02010086 OWR49188.1 JTDY01000422 KOB77258.1 BABH01004861 BABH01004862 NEVH01006731 PNF36811.1 KQ971343 EFA04151.1 PYGN01000830 PSN40265.1 GEZM01063934 JAV68904.1 KK852561 KDR21326.1 GECU01019935 JAS87771.1 GEBQ01009656 JAT30321.1 GECZ01009405 JAS60364.1 KZ288253 PBC31019.1 JR044938 AEY59920.1 QOIP01000009 RLU18516.1 KQ434777 KZC04247.1 LJIG01016254 KRT81153.1 KQ414578 KOC71205.1 NNAY01000193 OXU30116.1 GBYB01007072 JAG76839.1 GL440425 EFN65881.1 GEDC01021970 JAS15328.1 KK107038 EZA61934.1 GDKW01001278 JAI55317.1 GAHY01001286 JAA76224.1 GBGD01001361 JAC87528.1 GECL01003519 JAP02605.1 GFTR01006507 JAW09919.1 GBBI01000996 JAC17716.1 GBHO01035408 GBHO01035407 GBHO01030436 GBHO01030422 GBRD01002069 GBRD01002068 GDHC01015666 GDHC01012446 GDHC01003077 JAG08196.1 JAG08197.1 JAG13168.1 JAG13182.1 JAG63752.1 JAQ02963.1 JAQ06183.1 JAQ15552.1 AK417372 BAN20587.1 GDAI01000719 JAI16884.1 JRES01000438 KNC31324.1 KQ769580 OAD52854.1 GEMB01002826 JAS00373.1 KK854512 PTY16469.1 JXJN01025268 KA647557 AFP62186.1 CP012523 ALC38939.1 KB631904 ERL87078.1 CH902624 EDV33079.1 KPU74290.1 APGK01040727 KB740984 ENN76249.1 CH916368 EDW03701.1 CH933807 EDW13199.1 GAMC01012441 GAMC01012440 GAMC01012439 GAMC01012438 JAB94117.1 GDHF01009683 GDHF01009579 GDHF01009495 JAI42631.1 JAI42735.1 JAI42819.1 GAKP01007973 GAKP01007972 GAKP01007970 GAKP01007968 JAC50982.1 CH940649 EDW63809.1 DS235107 EEB12043.1 GFDF01007345 JAV06739.1 AXCM01001483 GBXI01015359 GBXI01001457 JAC98932.1 JAD12835.1 OUUW01000004 SPP79163.1 CH963857 EDW76144.1 CH479180 EDW29012.1 CH379060 EAL33785.1 GANO01004251 JAB55620.1 CM000157 EDW87711.1 KRJ97323.1 KRJ97322.1 AJWK01017307 GGFJ01004419 MBW53560.1 AF132185 AE014134 AAD34773.1 AAF52564.1 AAF52565.1 AGB92752.1 CH477313 EAT43857.1 GGFK01005364 MBW38685.1 CH954177 EDV59260.1 CM002910 KMY88863.1 CH480818 EDW52040.1 GGFK01005442 MBW38763.1 ADMH02000497 ETN66195.1 GGFM01003469 MBW24220.1 GFDL01008188 JAV26857.1 GFDL01008195 JAV26850.1 CM000361 EDX04137.1 APCN01000736 AAAB01008964 EAA12153.4 AXCN02000728
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000005204 UP000235965 UP000007266 UP000245037 UP000027135 UP000192223 UP000005203 UP000242457 UP000279307 UP000076502 UP000053825 UP000215335 UP000000311 UP000053097 UP000037069 UP000092445 UP000079169 UP000092443 UP000092460 UP000095301 UP000095300 UP000092553 UP000030742 UP000007801 UP000019118 UP000001070 UP000192221 UP000091820 UP000009192 UP000008792 UP000009046 UP000075883 UP000268350 UP000007798 UP000008744 UP000001819 UP000075920 UP000075882 UP000002282 UP000092461 UP000069272 UP000000803 UP000008820 UP000075881 UP000008711 UP000075900 UP000001292 UP000000673 UP000075902 UP000000304 UP000075884 UP000075840 UP000007062 UP000075886 UP000076407 UP000075903 UP000076408
UP000005204 UP000235965 UP000007266 UP000245037 UP000027135 UP000192223 UP000005203 UP000242457 UP000279307 UP000076502 UP000053825 UP000215335 UP000000311 UP000053097 UP000037069 UP000092445 UP000079169 UP000092443 UP000092460 UP000095301 UP000095300 UP000092553 UP000030742 UP000007801 UP000019118 UP000001070 UP000192221 UP000091820 UP000009192 UP000008792 UP000009046 UP000075883 UP000268350 UP000007798 UP000008744 UP000001819 UP000075920 UP000075882 UP000002282 UP000092461 UP000069272 UP000000803 UP000008820 UP000075881 UP000008711 UP000075900 UP000001292 UP000000673 UP000075902 UP000000304 UP000075884 UP000075840 UP000007062 UP000075886 UP000076407 UP000075903 UP000076408
Pfam
PF00481 PP2C
Interpro
SUPFAM
SSF81606
SSF81606
Gene 3D
CDD
ProteinModelPortal
A0A3S2NPY3
A0A2H1WGS4
A0A2A4K9H8
A0A1E1W2Z7
A0A194R337
A0A194PKU1
+ More
A0A212F632 A0A0L7LNZ4 H9J8Y3 A0A2J7R7K9 D6WLQ8 A0A2P8Y7P6 A0A1Y1L5Q3 A0A067RBQ9 A0A1B6ILH2 A0A1B6M308 A0A1W4X9I7 A0A1B6GD57 A0A088AQK8 A0A2A3EII8 V9IFR0 A0A3L8DDZ1 A0A154NX95 A0A0T6B1A8 A0A0L7RK12 A0A232FH79 A0A0C9R2K5 E2AKW4 A0A1B6CPH1 A0A026X3F9 A0A0P4VTL3 R4G3X5 A0A069DTV7 A0A0V0G3D6 A0A224XPJ5 A0A023F8K4 A0A0A9WXK6 R4WRP8 A0A0K8TR79 A0A0L0CIL8 A0A310S5L4 A0A161M4M2 A0A2R7WA99 A0A1B0AIU1 A0A3Q0J0B5 A0A1A9X8A1 A0A1B0C441 A0A1I8N0K0 T1PFI6 A0A1I8P0Q7 A0A0M3QTI3 U4U415 B3MV14 N6T7P8 B4JEI1 A0A1W4W8I8 A0A1A9WBW6 B4KG60 W8B689 A0A0K8VVC3 A0A034W9I4 B4LSS0 E0VF87 A0A1L8DJT2 A0A182MMC8 A0A0A1XPR0 A0A3B0JES6 B4MWK3 B4G9I2 Q29MA9 U5EEM8 A0A182VSH7 A0A182L024 B4NZG2 A0A0R1DJE4 A0A1B0CLI3 A0A2M4BKG9 A0A182FNB6 Q9XZ28 Q17C45 A0A182K1K7 A0A2M4AD20 B3N6P4 A0A182RWD2 A0A0J9QY22 B4HYB0 A0A2M4ADB2 W5JTB7 A0A2M3Z6V6 A0A1Q3FH28 A0A1Q3FH01 A0A182TMZ4 B4Q5U1 A0A182NT70 A0A182IBS9 Q7Q4Q8 A0A182QRH9 A0A182X198 A0A182UV48 A0A182YP54
A0A212F632 A0A0L7LNZ4 H9J8Y3 A0A2J7R7K9 D6WLQ8 A0A2P8Y7P6 A0A1Y1L5Q3 A0A067RBQ9 A0A1B6ILH2 A0A1B6M308 A0A1W4X9I7 A0A1B6GD57 A0A088AQK8 A0A2A3EII8 V9IFR0 A0A3L8DDZ1 A0A154NX95 A0A0T6B1A8 A0A0L7RK12 A0A232FH79 A0A0C9R2K5 E2AKW4 A0A1B6CPH1 A0A026X3F9 A0A0P4VTL3 R4G3X5 A0A069DTV7 A0A0V0G3D6 A0A224XPJ5 A0A023F8K4 A0A0A9WXK6 R4WRP8 A0A0K8TR79 A0A0L0CIL8 A0A310S5L4 A0A161M4M2 A0A2R7WA99 A0A1B0AIU1 A0A3Q0J0B5 A0A1A9X8A1 A0A1B0C441 A0A1I8N0K0 T1PFI6 A0A1I8P0Q7 A0A0M3QTI3 U4U415 B3MV14 N6T7P8 B4JEI1 A0A1W4W8I8 A0A1A9WBW6 B4KG60 W8B689 A0A0K8VVC3 A0A034W9I4 B4LSS0 E0VF87 A0A1L8DJT2 A0A182MMC8 A0A0A1XPR0 A0A3B0JES6 B4MWK3 B4G9I2 Q29MA9 U5EEM8 A0A182VSH7 A0A182L024 B4NZG2 A0A0R1DJE4 A0A1B0CLI3 A0A2M4BKG9 A0A182FNB6 Q9XZ28 Q17C45 A0A182K1K7 A0A2M4AD20 B3N6P4 A0A182RWD2 A0A0J9QY22 B4HYB0 A0A2M4ADB2 W5JTB7 A0A2M3Z6V6 A0A1Q3FH28 A0A1Q3FH01 A0A182TMZ4 B4Q5U1 A0A182NT70 A0A182IBS9 Q7Q4Q8 A0A182QRH9 A0A182X198 A0A182UV48 A0A182YP54
PDB
4OIC
E-value=2.82831e-24,
Score=278
Ontologies
GO
PANTHER
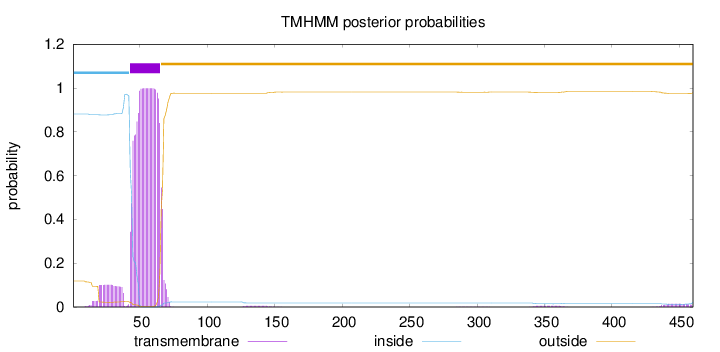
Topology
Length:
460
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.54738
Exp number, first 60 AAs:
17.86643
Total prob of N-in:
0.88108
POSSIBLE N-term signal
sequence
inside
1 - 42
TMhelix
43 - 65
outside
66 - 460
Population Genetic Test Statistics
Pi
318.288889
Theta
233.597831
Tajima's D
1.242965
CLR
0
CSRT
0.720113994300285
Interpretation
Uncertain
