Gene
KWMTBOMO01829 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005977
Annotation
C-type_lectin_5_precursor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.531
Sequence
CDS
ATGCAGGCGTGGTTGGTGGTGGCTTTCGTAGCTGCGTTGTCGGTTCTCGCGACCGCACAGAAGCCCGGTCGTTTCCTGTCGCTTCCCGTTGCGCATAAATGTGCTAACAGACCAAAGGAATTCTTTTACCAAGGACACAACTACTTCTACAGCGGTCACGTTCCCGAGCTAGCCAACCGGAAAGTAGACTGGTTGGACGGACGCAACATTTGCCGTGAATACTGCATGGACCTCGTCTCTATGGAGACCCAGGAAGAAAACAATCTGATCTTCAAGCTCATCCAGCAGAACGATGTTCCCTACATTTGGACATCGGGACGTCTATGCGACTTCAAGGGATGTGAAACGCGAAAAGATCTCGAACCCAAGAACATTCTCGGCTGGTTCTGGTCCGCTAACCGAGAAAAGATCTCGCCCACTAATCAGATACCGAACCGTTGGGGCTACAACCCTTGGTCTCAGACCGGGCACAAGAAGCAGCGCCAACCTGACAACGCAGAGTACGATATCAACGGTACTTCGGAGTCTTGTTTGAGCGTCCTCAACAACGTTTACAACGACGGCATCGCCTGGCACGACGTAGCCTGTTATCACGAGAAGCCCATCGTATGCGAAGACTCCGAGGAATTACTGAACTACATCGCGAGCAATAATCCCGGTCTCCGTCTATAA
Protein
MQAWLVVAFVAALSVLATAQKPGRFLSLPVAHKCANRPKEFFYQGHNYFYSGHVPELANRKVDWLDGRNICREYCMDLVSMETQEENNLIFKLIQQNDVPYIWTSGRLCDFKGCETRKDLEPKNILGWFWSANREKISPTNQIPNRWGYNPWSQTGHKKQRQPDNAEYDINGTSESCLSVLNNVYNDGIAWHDVACYHEKPIVCEDSEELLNYIASNNPGLRL
Summary
Uniprot
Q2F5Q5
H9J8Y5
A0A096ZGV8
A0A3S2TRN8
A0A212EN51
A0A0H4SZC6
+ More
I6RPF5 A0A2A4KA09 A0A194PS46 I4DLY5 I4DIH3 A0A219YXG4 D6WMQ9 A0A1W4XFW7 A0A1Y1KCI5 A0A195D4W9 A0A158NT65 A0A195EK58 K7ITK1 A0A232EQ47 N6UC69 B0WF13 A0A2H1WFI1 J3JVW0 A0A1B0CME0 A0A1Q3FPH0 A0A3L8DTE6 A0A310SN76 Q1HRD1 A0A182H9J5 A0A182N8K6 A0A023EIP1 A0A088AM42 A0A2A3E137 V9II58 A0A182YII5 A0A023ENM3 A0A182JCV2 A0A182PF77 A0A182R0I5 A0A182V1E8 A0A182KJP8 A0A182XB41 Q7Q0R9 A0A182R6T2 A0A182I0N6 E2B1M1 A0A084W641 A0A182LZC5 A0A182VT10 A0A2M4BYE5 A0A224XW22 T1DHR8 A0A182FQG1 T1H9B6 W5JHK1 A0A2M4AS58 A0A2M3ZE73 A0A0L7R4P7 A0A1D1Z3I4 A0A195F4X2 A0A154P861 A0A161MST6 A0A336L6G2 A0A146M885 A0A0A9XF86 E0VF89 A0A1B6LS38 A0A2K8JM51 A0A1B6LBN7 A0A1B6LFP1 A0A1V1FV79 B4KGY5 A0A2P8Y7L8 A0A0K8WGK6 A0A034W7C2 A0A0A1WQK2 B4LUA0 B4MTV9 A0A0M4DZU0 B3ML90 A0A1I8M961 A0A0L0CI81 A0A1I8PJJ0 A0A0J9QWM7 A0A0R1DJ38 B3N497 Q8SZK9 A0A067RE64 A0A1W4UW57 A0A3B0K941 B4I3C4 Q29LA7 B4GQC6 A0A1B0AIT4 B4NZ09 A0A1B0G4U4 A0A195BW36 F4WXS1 F2CR60
I6RPF5 A0A2A4KA09 A0A194PS46 I4DLY5 I4DIH3 A0A219YXG4 D6WMQ9 A0A1W4XFW7 A0A1Y1KCI5 A0A195D4W9 A0A158NT65 A0A195EK58 K7ITK1 A0A232EQ47 N6UC69 B0WF13 A0A2H1WFI1 J3JVW0 A0A1B0CME0 A0A1Q3FPH0 A0A3L8DTE6 A0A310SN76 Q1HRD1 A0A182H9J5 A0A182N8K6 A0A023EIP1 A0A088AM42 A0A2A3E137 V9II58 A0A182YII5 A0A023ENM3 A0A182JCV2 A0A182PF77 A0A182R0I5 A0A182V1E8 A0A182KJP8 A0A182XB41 Q7Q0R9 A0A182R6T2 A0A182I0N6 E2B1M1 A0A084W641 A0A182LZC5 A0A182VT10 A0A2M4BYE5 A0A224XW22 T1DHR8 A0A182FQG1 T1H9B6 W5JHK1 A0A2M4AS58 A0A2M3ZE73 A0A0L7R4P7 A0A1D1Z3I4 A0A195F4X2 A0A154P861 A0A161MST6 A0A336L6G2 A0A146M885 A0A0A9XF86 E0VF89 A0A1B6LS38 A0A2K8JM51 A0A1B6LBN7 A0A1B6LFP1 A0A1V1FV79 B4KGY5 A0A2P8Y7L8 A0A0K8WGK6 A0A034W7C2 A0A0A1WQK2 B4LUA0 B4MTV9 A0A0M4DZU0 B3ML90 A0A1I8M961 A0A0L0CI81 A0A1I8PJJ0 A0A0J9QWM7 A0A0R1DJ38 B3N497 Q8SZK9 A0A067RE64 A0A1W4UW57 A0A3B0K941 B4I3C4 Q29LA7 B4GQC6 A0A1B0AIT4 B4NZ09 A0A1B0G4U4 A0A195BW36 F4WXS1 F2CR60
Pubmed
19121390
22118469
26354079
22651552
18362917
19820115
+ More
28004739 21347285 20075255 28648823 23537049 22516182 30249741 17204158 17510324 26483478 24945155 25244985 20966253 12364791 14747013 17210077 20798317 24438588 20920257 23761445 26823975 25401762 20566863 28410430 17994087 29403074 25348373 25830018 25315136 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 15632085 21719571 21415278
28004739 21347285 20075255 28648823 23537049 22516182 30249741 17204158 17510324 26483478 24945155 25244985 20966253 12364791 14747013 17210077 20798317 24438588 20920257 23761445 26823975 25401762 20566863 28410430 17994087 29403074 25348373 25830018 25315136 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 15632085 21719571 21415278
EMBL
DQ311368
ABD36312.1
BABH01004853
KF752425
AIR96004.1
RSAL01000014
+ More
RVE53294.1 AGBW02013714 OWR42922.1 KP406778 AKP99432.1 JQ899036 AFM52345.1 NWSH01000024 PCG80623.1 KQ459601 KPI93950.1 AK402303 BAM18925.1 AK401091 BAM17713.1 KX550115 AQY54432.1 KQ971341 EFA04637.1 GEZM01090233 JAV57325.1 KQ976842 KYN07950.1 ADTU01025611 ADTU01025612 KQ978801 KYN28249.1 NNAY01002846 OXU20447.1 APGK01040724 APGK01040725 APGK01040726 KB740984 ENN76247.1 DS231913 EDS25921.1 ODYU01008337 SOQ51840.1 BT127378 AEE62340.1 AJWK01018611 GFDL01005679 JAV29366.1 QOIP01000005 RLU23169.1 KQ762903 OAD55298.1 DQ440163 CH477724 ABF18196.1 EAT36882.1 JXUM01004738 KQ560180 KXJ83981.1 GAPW01004290 JAC09308.1 KZ288467 PBC25407.1 JR046299 AEY60171.1 GAPW01003073 JAC10525.1 AXCN02001838 AAAB01008980 EAA14156.2 APCN01005224 GL444953 EFN60377.1 ATLV01020721 KE525305 KFB45685.1 AXCM01000242 GGFJ01008938 MBW58079.1 GFTR01003756 JAW12670.1 GAMD01002201 JAA99389.1 ACPB03001102 ADMH02001231 ETN63566.1 GGFK01010290 MBW43611.1 GGFM01005979 MBW26730.1 KQ414657 KOC65840.1 GDJX01006478 JAT61458.1 KQ981805 KYN35508.1 KQ434823 KZC07300.1 GEMB01000603 JAS02529.1 UFQS01002176 UFQT01002176 SSX13418.1 SSX32852.1 GDHC01020080 GDHC01002666 JAP98548.1 JAQ15963.1 GBHO01024207 GBHO01024123 GBHO01024116 GBHO01024097 GBHO01017359 GBHO01011735 GBHO01011734 GBHO01011730 JAG19397.1 JAG19481.1 JAG19488.1 JAG19507.1 JAG26245.1 JAG31869.1 JAG31870.1 JAG31874.1 DS235107 EEB12045.1 GEBQ01013512 JAT26465.1 KY031157 ATU82908.1 GEBQ01018878 JAT21099.1 GEBQ01017464 JAT22513.1 FX985255 BAX07268.1 CH933807 EDW11185.1 PYGN01000830 PSN40258.1 GDHF01002110 JAI50204.1 GAKP01008343 JAC50609.1 GBXI01013140 JAD01152.1 CH940649 EDW64087.1 CH963852 EDW75548.2 CP012523 ALC38191.1 ALC39985.1 CH902620 EDV31708.1 JRES01000438 KNC31204.1 CM002910 KMY88119.1 CM000157 KRJ97291.1 CH954177 EDV57767.1 AE014134 AY070683 BT014659 AAF50981.4 AAL48154.1 AAT27283.1 AGB92582.1 KK852561 KDR21328.1 OUUW01000004 SPP80048.1 CH480820 EDW54269.1 CH379060 EAL34138.3 CH479187 EDW39798.1 EDW87673.2 CCAG010022902 KQ976403 KYM92176.1 GL888427 EGI60999.1 AK354112 BAJ85331.1
RVE53294.1 AGBW02013714 OWR42922.1 KP406778 AKP99432.1 JQ899036 AFM52345.1 NWSH01000024 PCG80623.1 KQ459601 KPI93950.1 AK402303 BAM18925.1 AK401091 BAM17713.1 KX550115 AQY54432.1 KQ971341 EFA04637.1 GEZM01090233 JAV57325.1 KQ976842 KYN07950.1 ADTU01025611 ADTU01025612 KQ978801 KYN28249.1 NNAY01002846 OXU20447.1 APGK01040724 APGK01040725 APGK01040726 KB740984 ENN76247.1 DS231913 EDS25921.1 ODYU01008337 SOQ51840.1 BT127378 AEE62340.1 AJWK01018611 GFDL01005679 JAV29366.1 QOIP01000005 RLU23169.1 KQ762903 OAD55298.1 DQ440163 CH477724 ABF18196.1 EAT36882.1 JXUM01004738 KQ560180 KXJ83981.1 GAPW01004290 JAC09308.1 KZ288467 PBC25407.1 JR046299 AEY60171.1 GAPW01003073 JAC10525.1 AXCN02001838 AAAB01008980 EAA14156.2 APCN01005224 GL444953 EFN60377.1 ATLV01020721 KE525305 KFB45685.1 AXCM01000242 GGFJ01008938 MBW58079.1 GFTR01003756 JAW12670.1 GAMD01002201 JAA99389.1 ACPB03001102 ADMH02001231 ETN63566.1 GGFK01010290 MBW43611.1 GGFM01005979 MBW26730.1 KQ414657 KOC65840.1 GDJX01006478 JAT61458.1 KQ981805 KYN35508.1 KQ434823 KZC07300.1 GEMB01000603 JAS02529.1 UFQS01002176 UFQT01002176 SSX13418.1 SSX32852.1 GDHC01020080 GDHC01002666 JAP98548.1 JAQ15963.1 GBHO01024207 GBHO01024123 GBHO01024116 GBHO01024097 GBHO01017359 GBHO01011735 GBHO01011734 GBHO01011730 JAG19397.1 JAG19481.1 JAG19488.1 JAG19507.1 JAG26245.1 JAG31869.1 JAG31870.1 JAG31874.1 DS235107 EEB12045.1 GEBQ01013512 JAT26465.1 KY031157 ATU82908.1 GEBQ01018878 JAT21099.1 GEBQ01017464 JAT22513.1 FX985255 BAX07268.1 CH933807 EDW11185.1 PYGN01000830 PSN40258.1 GDHF01002110 JAI50204.1 GAKP01008343 JAC50609.1 GBXI01013140 JAD01152.1 CH940649 EDW64087.1 CH963852 EDW75548.2 CP012523 ALC38191.1 ALC39985.1 CH902620 EDV31708.1 JRES01000438 KNC31204.1 CM002910 KMY88119.1 CM000157 KRJ97291.1 CH954177 EDV57767.1 AE014134 AY070683 BT014659 AAF50981.4 AAL48154.1 AAT27283.1 AGB92582.1 KK852561 KDR21328.1 OUUW01000004 SPP80048.1 CH480820 EDW54269.1 CH379060 EAL34138.3 CH479187 EDW39798.1 EDW87673.2 CCAG010022902 KQ976403 KYM92176.1 GL888427 EGI60999.1 AK354112 BAJ85331.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000218220
UP000053268
UP000007266
+ More
UP000192223 UP000078542 UP000005205 UP000078492 UP000002358 UP000215335 UP000019118 UP000002320 UP000092461 UP000279307 UP000008820 UP000069940 UP000249989 UP000075884 UP000005203 UP000242457 UP000076408 UP000075880 UP000075885 UP000075886 UP000075903 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840 UP000000311 UP000030765 UP000075883 UP000075920 UP000069272 UP000015103 UP000000673 UP000053825 UP000078541 UP000076502 UP000009046 UP000009192 UP000245037 UP000008792 UP000007798 UP000092553 UP000007801 UP000095301 UP000037069 UP000095300 UP000002282 UP000008711 UP000000803 UP000027135 UP000192221 UP000268350 UP000001292 UP000001819 UP000008744 UP000092445 UP000092444 UP000078540 UP000007755
UP000192223 UP000078542 UP000005205 UP000078492 UP000002358 UP000215335 UP000019118 UP000002320 UP000092461 UP000279307 UP000008820 UP000069940 UP000249989 UP000075884 UP000005203 UP000242457 UP000076408 UP000075880 UP000075885 UP000075886 UP000075903 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840 UP000000311 UP000030765 UP000075883 UP000075920 UP000069272 UP000015103 UP000000673 UP000053825 UP000078541 UP000076502 UP000009046 UP000009192 UP000245037 UP000008792 UP000007798 UP000092553 UP000007801 UP000095301 UP000037069 UP000095300 UP000002282 UP000008711 UP000000803 UP000027135 UP000192221 UP000268350 UP000001292 UP000001819 UP000008744 UP000092445 UP000092444 UP000078540 UP000007755
Pfam
PF00059 Lectin_C
Interpro
SUPFAM
SSF56436
SSF56436
Gene 3D
ProteinModelPortal
Q2F5Q5
H9J8Y5
A0A096ZGV8
A0A3S2TRN8
A0A212EN51
A0A0H4SZC6
+ More
I6RPF5 A0A2A4KA09 A0A194PS46 I4DLY5 I4DIH3 A0A219YXG4 D6WMQ9 A0A1W4XFW7 A0A1Y1KCI5 A0A195D4W9 A0A158NT65 A0A195EK58 K7ITK1 A0A232EQ47 N6UC69 B0WF13 A0A2H1WFI1 J3JVW0 A0A1B0CME0 A0A1Q3FPH0 A0A3L8DTE6 A0A310SN76 Q1HRD1 A0A182H9J5 A0A182N8K6 A0A023EIP1 A0A088AM42 A0A2A3E137 V9II58 A0A182YII5 A0A023ENM3 A0A182JCV2 A0A182PF77 A0A182R0I5 A0A182V1E8 A0A182KJP8 A0A182XB41 Q7Q0R9 A0A182R6T2 A0A182I0N6 E2B1M1 A0A084W641 A0A182LZC5 A0A182VT10 A0A2M4BYE5 A0A224XW22 T1DHR8 A0A182FQG1 T1H9B6 W5JHK1 A0A2M4AS58 A0A2M3ZE73 A0A0L7R4P7 A0A1D1Z3I4 A0A195F4X2 A0A154P861 A0A161MST6 A0A336L6G2 A0A146M885 A0A0A9XF86 E0VF89 A0A1B6LS38 A0A2K8JM51 A0A1B6LBN7 A0A1B6LFP1 A0A1V1FV79 B4KGY5 A0A2P8Y7L8 A0A0K8WGK6 A0A034W7C2 A0A0A1WQK2 B4LUA0 B4MTV9 A0A0M4DZU0 B3ML90 A0A1I8M961 A0A0L0CI81 A0A1I8PJJ0 A0A0J9QWM7 A0A0R1DJ38 B3N497 Q8SZK9 A0A067RE64 A0A1W4UW57 A0A3B0K941 B4I3C4 Q29LA7 B4GQC6 A0A1B0AIT4 B4NZ09 A0A1B0G4U4 A0A195BW36 F4WXS1 F2CR60
I6RPF5 A0A2A4KA09 A0A194PS46 I4DLY5 I4DIH3 A0A219YXG4 D6WMQ9 A0A1W4XFW7 A0A1Y1KCI5 A0A195D4W9 A0A158NT65 A0A195EK58 K7ITK1 A0A232EQ47 N6UC69 B0WF13 A0A2H1WFI1 J3JVW0 A0A1B0CME0 A0A1Q3FPH0 A0A3L8DTE6 A0A310SN76 Q1HRD1 A0A182H9J5 A0A182N8K6 A0A023EIP1 A0A088AM42 A0A2A3E137 V9II58 A0A182YII5 A0A023ENM3 A0A182JCV2 A0A182PF77 A0A182R0I5 A0A182V1E8 A0A182KJP8 A0A182XB41 Q7Q0R9 A0A182R6T2 A0A182I0N6 E2B1M1 A0A084W641 A0A182LZC5 A0A182VT10 A0A2M4BYE5 A0A224XW22 T1DHR8 A0A182FQG1 T1H9B6 W5JHK1 A0A2M4AS58 A0A2M3ZE73 A0A0L7R4P7 A0A1D1Z3I4 A0A195F4X2 A0A154P861 A0A161MST6 A0A336L6G2 A0A146M885 A0A0A9XF86 E0VF89 A0A1B6LS38 A0A2K8JM51 A0A1B6LBN7 A0A1B6LFP1 A0A1V1FV79 B4KGY5 A0A2P8Y7L8 A0A0K8WGK6 A0A034W7C2 A0A0A1WQK2 B4LUA0 B4MTV9 A0A0M4DZU0 B3ML90 A0A1I8M961 A0A0L0CI81 A0A1I8PJJ0 A0A0J9QWM7 A0A0R1DJ38 B3N497 Q8SZK9 A0A067RE64 A0A1W4UW57 A0A3B0K941 B4I3C4 Q29LA7 B4GQC6 A0A1B0AIT4 B4NZ09 A0A1B0G4U4 A0A195BW36 F4WXS1 F2CR60
Ontologies
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.996298
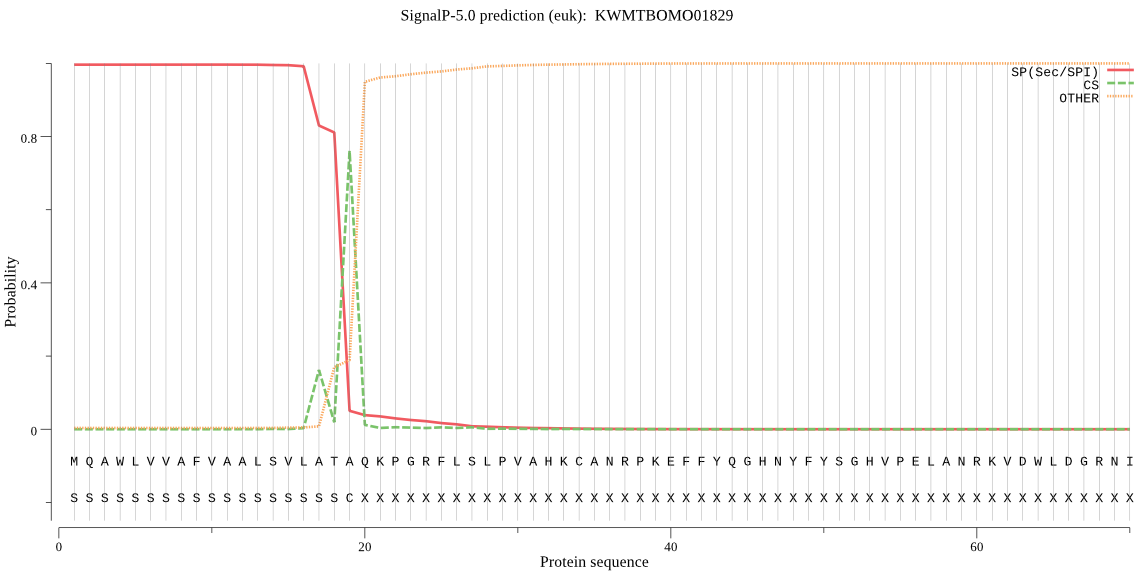
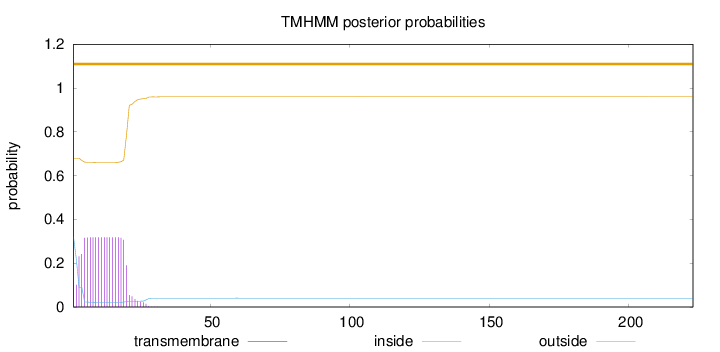
Length:
223
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.75146
Exp number, first 60 AAs:
5.75146
Total prob of N-in:
0.32037
outside
1 - 223
Population Genetic Test Statistics
Pi
23.444775
Theta
186.560149
Tajima's D
0.561017
CLR
0.636001
CSRT
0.530123493825309
Interpretation
Uncertain
