Pre Gene Modal
BGIBMGA006173
Annotation
transcription_elongation_factor_S-II_[Bombyx_mori]
Full name
Transcription elongation factor S-II
Alternative Name
RNA polymerase II elongation factor DMS-II
TFIIS
TFIIS
Location in the cell
PlasmaMembrane Reliability : 4.875
Sequence
CDS
ATGAGCGTTGAAGAAGATGTTATGAAAATACAGAAAAAGCTTACTAAAATGACATCTGAAGACGGCACGGGTCAAGAAGAAGCGTTAGAACTTCTGAAAGCACTTCAAACAATGGCTATCAACTTGGACGTCTTAACGAAAACAAGAATAGGAATGACTGTCAATGCTCTGAGAAAATCTAGCAAAGATGATGAAGTTATATCGCTTTGTAAAACACTAATTAAAAACTGGAAAAAGTTTTTGTCGACACCAAACACGACTAAGGACAATGGAAATTCATCCAAATCTAAAAAAGAAGGTAAAGACAAAGATAAAAAAGAAGATAGAGATAAGAAATTGCCAGCATCATTTCCACCACAGTCCAACACTACAGATGCAGTAAGGCTCAAGTGTAGAGAACTTCTAACCCAAGCACTCAAGGCTGCTGGTGAAACTTCCAATGCTTGCGGATCTCCTGAAGAACTTGCTGAAGAGTTAGAAGAGTGTATTTATGCTGAATTCAAAAACACTGACATGCGTTACAAAAACAGGGTTAGATCAAGAGTTGCTAACTTGAAAGATCCTAAGAACCCAACATTACGTACAAATTTCTTTAATGGTGTCATTTCAGCATCACGTTTAGCTAAAATGACTCCTGAAGAAATGGCAAGTGATGAAATGAAAAAGTTGAGAGAAAAGTTCATTAAAGAAGCTATTGATGATGCACAGCTTGCCACAGTTCAAGGCACCAAAACAGAAATGTTAAAATGTGGCAAATGCAAAAAGAAGAACTGTACCTACAATCAACTACAAACAAGAAGTTCCGATGAACCTATGACCACATTTGTCCTTTGTAATGAATGTGGTAATCGCTGGAAATTTTGCTAA
Protein
MSVEEDVMKIQKKLTKMTSEDGTGQEEALELLKALQTMAINLDVLTKTRIGMTVNALRKSSKDDEVISLCKTLIKNWKKFLSTPNTTKDNGNSSKSKKEGKDKDKKEDRDKKLPASFPPQSNTTDAVRLKCRELLTQALKAAGETSNACGSPEELAEELEECIYAEFKNTDMRYKNRVRSRVANLKDPKNPTLRTNFFNGVISASRLAKMTPEEMASDEMKKLREKFIKEAIDDAQLATVQGTKTEMLKCGKCKKKNCTYNQLQTRSSDEPMTTFVLCNECGNRWKFC
Summary
Miscellaneous
S-II binds to RNA-polymerase II in the absence of transcription.
Similarity
Belongs to the TFS-II family.
Keywords
Complete proteome
DNA-binding
Metal-binding
Nucleus
Reference proteome
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Transcription elongation factor S-II
Uniprot
Q1HQ13
A0A194R2B3
A0A212EN69
A0A194PMJ0
A0A2H1WPL0
A0A3S2LT57
+ More
A0A2A4K974 S4PMU1 A0A0L7LE19 A0A2J7QYY3 A0A067R679 D6WJB1 A0A1W4W5Z2 A0A0K8TRH6 A0A1B6CUU0 Q17L48 A0A1S4EYX3 A0A1Y1L765 A0A1L8DH18 A0A023EMX9 A0A0T6AVA4 T1DGQ7 A0A182G2J9 A0A1B0GNB8 T1E273 B3MMA2 A0A0M5J1S9 A0A1I8NPU9 Q29P72 B4GK82 A0A1Q3EU47 A0A0J9R248 M9PFV9 P20232 B4HXM1 A0A0N7Z9H5 B3N5N7 A0A3B0K725 A0A023F953 A0A336M7E8 A0A0V0GA48 A0A224XPJ8 A0A0C9R8I7 A0A069DS80 A0A084VYE3 A0A026WIF4 A0A1Q3FDY2 A0A158NPH3 A0A195BA67 E2ACU5 A0A336LVW6 A0A0J7L8V4 A0A1Q3G1J1 A0A2A3EJQ6 V9IF07 A0A088A1K6 R4WSV9 U5EY68 K7JA93 A0A1S4JDE4 A0A0A9YCX9 T1HP11 B0WC04 A0A182G2K0 A0A195EUZ9 B0WC05 F4WNZ9 A0A154PMF1 A0A195E627 A0A2H8TYJ1 A0A2S2NC30 E0VT44 J9JYJ3 A0A1V9XQZ1 N6U6L7 A0A2H8TP85 R4FPX7 A0A1B6LS96 A0A0P5U9W5 A0A195CN06 A0A0P5ZVC4 A0A0P5V671 A0A0P5E2D0 A0A0P4Y4K2 E9GR89 A0A0P6HLK0 A0A151WRA7 A0A0N8CEG8 A0A0P5TTR6 A0A3S1BGD6 A0A293MEV4 T1J687 A0A0B7A7F5 A0A1D2MLU8 A0A2C9JR05 A0A2R5LL96 A0A226F1N5 A0A131XQP3 A0A023FV83 U4UGQ9
A0A2A4K974 S4PMU1 A0A0L7LE19 A0A2J7QYY3 A0A067R679 D6WJB1 A0A1W4W5Z2 A0A0K8TRH6 A0A1B6CUU0 Q17L48 A0A1S4EYX3 A0A1Y1L765 A0A1L8DH18 A0A023EMX9 A0A0T6AVA4 T1DGQ7 A0A182G2J9 A0A1B0GNB8 T1E273 B3MMA2 A0A0M5J1S9 A0A1I8NPU9 Q29P72 B4GK82 A0A1Q3EU47 A0A0J9R248 M9PFV9 P20232 B4HXM1 A0A0N7Z9H5 B3N5N7 A0A3B0K725 A0A023F953 A0A336M7E8 A0A0V0GA48 A0A224XPJ8 A0A0C9R8I7 A0A069DS80 A0A084VYE3 A0A026WIF4 A0A1Q3FDY2 A0A158NPH3 A0A195BA67 E2ACU5 A0A336LVW6 A0A0J7L8V4 A0A1Q3G1J1 A0A2A3EJQ6 V9IF07 A0A088A1K6 R4WSV9 U5EY68 K7JA93 A0A1S4JDE4 A0A0A9YCX9 T1HP11 B0WC04 A0A182G2K0 A0A195EUZ9 B0WC05 F4WNZ9 A0A154PMF1 A0A195E627 A0A2H8TYJ1 A0A2S2NC30 E0VT44 J9JYJ3 A0A1V9XQZ1 N6U6L7 A0A2H8TP85 R4FPX7 A0A1B6LS96 A0A0P5U9W5 A0A195CN06 A0A0P5ZVC4 A0A0P5V671 A0A0P5E2D0 A0A0P4Y4K2 E9GR89 A0A0P6HLK0 A0A151WRA7 A0A0N8CEG8 A0A0P5TTR6 A0A3S1BGD6 A0A293MEV4 T1J687 A0A0B7A7F5 A0A1D2MLU8 A0A2C9JR05 A0A2R5LL96 A0A226F1N5 A0A131XQP3 A0A023FV83 U4UGQ9
Pubmed
19121390
26354079
22118469
23622113
26227816
24845553
+ More
18362917 19820115 26369729 17510324 28004739 24945155 24330624 26483478 17994087 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2243775 7772609 10471707 12537569 27129103 25474469 26334808 24438588 24508170 30249741 21347285 20798317 23691247 20075255 25401762 26823975 21719571 20566863 28327890 23537049 21292972 27289101 15562597 28049606
18362917 19820115 26369729 17510324 28004739 24945155 24330624 26483478 17994087 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2243775 7772609 10471707 12537569 27129103 25474469 26334808 24438588 24508170 30249741 21347285 20798317 23691247 20075255 25401762 26823975 21719571 20566863 28327890 23537049 21292972 27289101 15562597 28049606
EMBL
BABH01004853
DQ443239
ABF51328.1
KQ460855
KPJ11928.1
AGBW02013714
+ More
OWR42926.1 KQ459601 KPI93954.1 ODYU01010122 SOQ55010.1 RSAL01000014 RVE53297.1 NWSH01000024 PCG80619.1 GAIX01000001 JAA92559.1 JTDY01001585 KOB73451.1 NEVH01009082 PNF33794.1 KK852860 KDR14833.1 KQ971343 EFA04452.1 GDAI01000865 JAI16738.1 GEDC01020056 JAS17242.1 CH477219 EAT47378.1 GEZM01068531 JAV66907.1 GFDF01008407 JAV05677.1 GAPW01002865 JAC10733.1 LJIG01022774 KRT78789.1 GALA01001566 JAA93286.1 JXUM01040106 KQ561232 KXJ79286.1 AJVK01013111 GALA01000994 JAA93858.1 CH902620 EDV31862.1 CP012523 ALC39718.1 CH379058 EAL34421.1 CH479184 EDW37048.1 GFDL01016218 JAV18827.1 CM002910 KMY90327.1 AE014134 AGB92992.1 X53670 L26091 AY051843 CH480818 EDW51801.1 GDKW01000531 JAI56064.1 CH954177 EDV57996.1 OUUW01000046 SPP90007.1 GBBI01000690 JAC18022.1 UFQT01000634 SSX25950.1 GECL01001853 JAP04271.1 GFTR01004668 JAW11758.1 GBYB01004405 GBYB01004407 GBYB01004409 GBYB01008919 JAG74172.1 JAG74174.1 JAG74176.1 JAG78686.1 GBGD01002377 JAC86512.1 ATLV01018360 ATLV01018361 KE525231 KFB42987.1 KK107207 QOIP01000002 EZA55441.1 RLU25535.1 GFDL01009272 JAV25773.1 ADTU01022546 KQ976542 KYM81089.1 GL438582 EFN68743.1 UFQS01000222 UFQT01000222 SSX01580.1 SSX21960.1 LBMM01000257 KMR02736.1 GFDL01001389 JAV33656.1 KZ288241 PBC31271.1 JR042625 AEY59645.1 AK417802 BAN21017.1 GANO01000490 JAB59381.1 AAZX01002936 AAZX01004281 AAZX01004313 AAZX01008587 AAZX01012701 AAZX01014769 AAZX01017293 GBHO01014133 GBRD01015541 GDHC01011969 JAG29471.1 JAG50285.1 JAQ06660.1 ACPB03014845 DS231884 EDS43064.1 JXUM01040107 KXJ79287.1 KQ981965 KYN31709.1 EDS43065.1 GL888243 EGI63930.1 KQ434978 KZC13032.1 KQ979608 KYN20337.1 GFXV01006413 MBW18218.1 GGMR01002049 MBY14668.1 DS235759 EEB16550.1 ABLF02035743 MNPL01005592 OQR75926.1 APGK01037237 KB740948 ENN77285.1 GFXV01004005 MBW15810.1 GAHY01000053 JAA77457.1 GEBQ01013396 JAT26581.1 GDIP01119754 JAL83960.1 KQ977574 KYN01847.1 GDIP01051510 JAM52205.1 GDIP01119753 JAL83961.1 GDIP01147860 GDIQ01224916 JAJ75542.1 JAK26809.1 GDIP01233840 JAI89561.1 GL732559 EFX77969.1 GDIQ01017062 JAN77675.1 KQ982807 KYQ50396.1 GDIP01139703 JAL64011.1 GDIP01139702 JAL64012.1 RQTK01000247 RUS83339.1 GFWV01019519 MAA44247.1 JH431873 HACG01029035 CEK75900.1 LJIJ01000880 ODM93973.1 GGLE01006174 MBY10300.1 LNIX01000001 OXA63334.1 GEFH01000575 JAP68006.1 GBBL01001554 JAC25766.1 KB632348 ERL93164.1
OWR42926.1 KQ459601 KPI93954.1 ODYU01010122 SOQ55010.1 RSAL01000014 RVE53297.1 NWSH01000024 PCG80619.1 GAIX01000001 JAA92559.1 JTDY01001585 KOB73451.1 NEVH01009082 PNF33794.1 KK852860 KDR14833.1 KQ971343 EFA04452.1 GDAI01000865 JAI16738.1 GEDC01020056 JAS17242.1 CH477219 EAT47378.1 GEZM01068531 JAV66907.1 GFDF01008407 JAV05677.1 GAPW01002865 JAC10733.1 LJIG01022774 KRT78789.1 GALA01001566 JAA93286.1 JXUM01040106 KQ561232 KXJ79286.1 AJVK01013111 GALA01000994 JAA93858.1 CH902620 EDV31862.1 CP012523 ALC39718.1 CH379058 EAL34421.1 CH479184 EDW37048.1 GFDL01016218 JAV18827.1 CM002910 KMY90327.1 AE014134 AGB92992.1 X53670 L26091 AY051843 CH480818 EDW51801.1 GDKW01000531 JAI56064.1 CH954177 EDV57996.1 OUUW01000046 SPP90007.1 GBBI01000690 JAC18022.1 UFQT01000634 SSX25950.1 GECL01001853 JAP04271.1 GFTR01004668 JAW11758.1 GBYB01004405 GBYB01004407 GBYB01004409 GBYB01008919 JAG74172.1 JAG74174.1 JAG74176.1 JAG78686.1 GBGD01002377 JAC86512.1 ATLV01018360 ATLV01018361 KE525231 KFB42987.1 KK107207 QOIP01000002 EZA55441.1 RLU25535.1 GFDL01009272 JAV25773.1 ADTU01022546 KQ976542 KYM81089.1 GL438582 EFN68743.1 UFQS01000222 UFQT01000222 SSX01580.1 SSX21960.1 LBMM01000257 KMR02736.1 GFDL01001389 JAV33656.1 KZ288241 PBC31271.1 JR042625 AEY59645.1 AK417802 BAN21017.1 GANO01000490 JAB59381.1 AAZX01002936 AAZX01004281 AAZX01004313 AAZX01008587 AAZX01012701 AAZX01014769 AAZX01017293 GBHO01014133 GBRD01015541 GDHC01011969 JAG29471.1 JAG50285.1 JAQ06660.1 ACPB03014845 DS231884 EDS43064.1 JXUM01040107 KXJ79287.1 KQ981965 KYN31709.1 EDS43065.1 GL888243 EGI63930.1 KQ434978 KZC13032.1 KQ979608 KYN20337.1 GFXV01006413 MBW18218.1 GGMR01002049 MBY14668.1 DS235759 EEB16550.1 ABLF02035743 MNPL01005592 OQR75926.1 APGK01037237 KB740948 ENN77285.1 GFXV01004005 MBW15810.1 GAHY01000053 JAA77457.1 GEBQ01013396 JAT26581.1 GDIP01119754 JAL83960.1 KQ977574 KYN01847.1 GDIP01051510 JAM52205.1 GDIP01119753 JAL83961.1 GDIP01147860 GDIQ01224916 JAJ75542.1 JAK26809.1 GDIP01233840 JAI89561.1 GL732559 EFX77969.1 GDIQ01017062 JAN77675.1 KQ982807 KYQ50396.1 GDIP01139703 JAL64011.1 GDIP01139702 JAL64012.1 RQTK01000247 RUS83339.1 GFWV01019519 MAA44247.1 JH431873 HACG01029035 CEK75900.1 LJIJ01000880 ODM93973.1 GGLE01006174 MBY10300.1 LNIX01000001 OXA63334.1 GEFH01000575 JAP68006.1 GBBL01001554 JAC25766.1 KB632348 ERL93164.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000283053
UP000218220
+ More
UP000037510 UP000235965 UP000027135 UP000007266 UP000192223 UP000008820 UP000069940 UP000249989 UP000092462 UP000007801 UP000092553 UP000095300 UP000001819 UP000008744 UP000000803 UP000001292 UP000008711 UP000268350 UP000030765 UP000053097 UP000279307 UP000005205 UP000078540 UP000000311 UP000036403 UP000242457 UP000005203 UP000002358 UP000015103 UP000002320 UP000078541 UP000007755 UP000076502 UP000078492 UP000009046 UP000007819 UP000192247 UP000019118 UP000078542 UP000000305 UP000075809 UP000271974 UP000094527 UP000076420 UP000198287 UP000030742
UP000037510 UP000235965 UP000027135 UP000007266 UP000192223 UP000008820 UP000069940 UP000249989 UP000092462 UP000007801 UP000092553 UP000095300 UP000001819 UP000008744 UP000000803 UP000001292 UP000008711 UP000268350 UP000030765 UP000053097 UP000279307 UP000005205 UP000078540 UP000000311 UP000036403 UP000242457 UP000005203 UP000002358 UP000015103 UP000002320 UP000078541 UP000007755 UP000076502 UP000078492 UP000009046 UP000007819 UP000192247 UP000019118 UP000078542 UP000000305 UP000075809 UP000271974 UP000094527 UP000076420 UP000198287 UP000030742
Interpro
Gene 3D
ProteinModelPortal
Q1HQ13
A0A194R2B3
A0A212EN69
A0A194PMJ0
A0A2H1WPL0
A0A3S2LT57
+ More
A0A2A4K974 S4PMU1 A0A0L7LE19 A0A2J7QYY3 A0A067R679 D6WJB1 A0A1W4W5Z2 A0A0K8TRH6 A0A1B6CUU0 Q17L48 A0A1S4EYX3 A0A1Y1L765 A0A1L8DH18 A0A023EMX9 A0A0T6AVA4 T1DGQ7 A0A182G2J9 A0A1B0GNB8 T1E273 B3MMA2 A0A0M5J1S9 A0A1I8NPU9 Q29P72 B4GK82 A0A1Q3EU47 A0A0J9R248 M9PFV9 P20232 B4HXM1 A0A0N7Z9H5 B3N5N7 A0A3B0K725 A0A023F953 A0A336M7E8 A0A0V0GA48 A0A224XPJ8 A0A0C9R8I7 A0A069DS80 A0A084VYE3 A0A026WIF4 A0A1Q3FDY2 A0A158NPH3 A0A195BA67 E2ACU5 A0A336LVW6 A0A0J7L8V4 A0A1Q3G1J1 A0A2A3EJQ6 V9IF07 A0A088A1K6 R4WSV9 U5EY68 K7JA93 A0A1S4JDE4 A0A0A9YCX9 T1HP11 B0WC04 A0A182G2K0 A0A195EUZ9 B0WC05 F4WNZ9 A0A154PMF1 A0A195E627 A0A2H8TYJ1 A0A2S2NC30 E0VT44 J9JYJ3 A0A1V9XQZ1 N6U6L7 A0A2H8TP85 R4FPX7 A0A1B6LS96 A0A0P5U9W5 A0A195CN06 A0A0P5ZVC4 A0A0P5V671 A0A0P5E2D0 A0A0P4Y4K2 E9GR89 A0A0P6HLK0 A0A151WRA7 A0A0N8CEG8 A0A0P5TTR6 A0A3S1BGD6 A0A293MEV4 T1J687 A0A0B7A7F5 A0A1D2MLU8 A0A2C9JR05 A0A2R5LL96 A0A226F1N5 A0A131XQP3 A0A023FV83 U4UGQ9
A0A2A4K974 S4PMU1 A0A0L7LE19 A0A2J7QYY3 A0A067R679 D6WJB1 A0A1W4W5Z2 A0A0K8TRH6 A0A1B6CUU0 Q17L48 A0A1S4EYX3 A0A1Y1L765 A0A1L8DH18 A0A023EMX9 A0A0T6AVA4 T1DGQ7 A0A182G2J9 A0A1B0GNB8 T1E273 B3MMA2 A0A0M5J1S9 A0A1I8NPU9 Q29P72 B4GK82 A0A1Q3EU47 A0A0J9R248 M9PFV9 P20232 B4HXM1 A0A0N7Z9H5 B3N5N7 A0A3B0K725 A0A023F953 A0A336M7E8 A0A0V0GA48 A0A224XPJ8 A0A0C9R8I7 A0A069DS80 A0A084VYE3 A0A026WIF4 A0A1Q3FDY2 A0A158NPH3 A0A195BA67 E2ACU5 A0A336LVW6 A0A0J7L8V4 A0A1Q3G1J1 A0A2A3EJQ6 V9IF07 A0A088A1K6 R4WSV9 U5EY68 K7JA93 A0A1S4JDE4 A0A0A9YCX9 T1HP11 B0WC04 A0A182G2K0 A0A195EUZ9 B0WC05 F4WNZ9 A0A154PMF1 A0A195E627 A0A2H8TYJ1 A0A2S2NC30 E0VT44 J9JYJ3 A0A1V9XQZ1 N6U6L7 A0A2H8TP85 R4FPX7 A0A1B6LS96 A0A0P5U9W5 A0A195CN06 A0A0P5ZVC4 A0A0P5V671 A0A0P5E2D0 A0A0P4Y4K2 E9GR89 A0A0P6HLK0 A0A151WRA7 A0A0N8CEG8 A0A0P5TTR6 A0A3S1BGD6 A0A293MEV4 T1J687 A0A0B7A7F5 A0A1D2MLU8 A0A2C9JR05 A0A2R5LL96 A0A226F1N5 A0A131XQP3 A0A023FV83 U4UGQ9
PDB
6O9L
E-value=1.69591e-58,
Score=571
Ontologies
KEGG
GO
Topology
Subcellular location
Nucleus
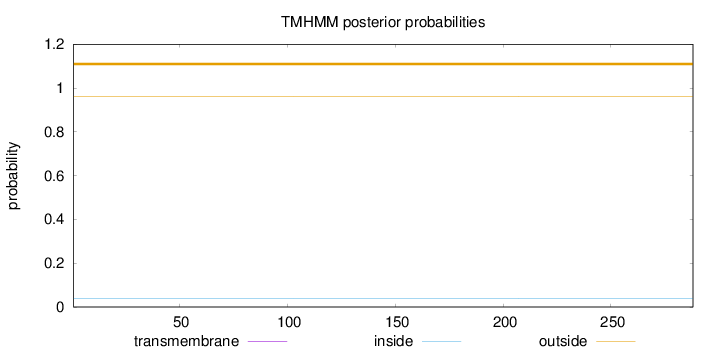
Length:
288
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0004
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.03932
outside
1 - 288
Population Genetic Test Statistics
Pi
235.263938
Theta
193.25756
Tajima's D
0.791095
CLR
0.459789
CSRT
0.595020248987551
Interpretation
Uncertain
