Gene
KWMTBOMO01821 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006171
Annotation
PREDICTED:_fibronectin_type-III_domain-containing_protein_3a_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.44 Nuclear Reliability : 1.252 PlasmaMembrane Reliability : 1.155
Sequence
CDS
ATGGTTGGCGTGGGCGTCGCGGAGGCAGGGGGTGGGGGAGGCGGCGTCCCCGGCGACGGCTACTATGGGGAGTACTACCCTCCGGAGCAATACTACGTACCCCCGGACATGTGTCCTCATCCACAGCAACATCCGCAACATGCACACATGTGCACTGTGTTACCTGAATACGGCGGCCTACCAGTGGTTACGTCGGGCACCATGATGCCGCCTTTGATGCCACCAGTGCTCGACGAGAACATGCGCCAATATTTGATGCACCAGCACGCTCAACACCCGCATCACGCGCCGCATCATCAACCGCCGCACCACCAGCCCCCGCATCATCAGCCGCCTCCGCATCACCAACCTCCTCATCATTATGGTCCAACCAATGGTGCTGGTGGTCCACAGCATTTCTATGGTGCGGGGTACCCAGCCCATTTTCACCATGTTCCCCCACACCATATGCAGCATTCGCCGCCACCGCCTATATACCAGAAGGATGAGCGAACTCAGAGGCAGTACTCCAAGTTAAAACAGAAGTTAGAAAGAAAAAACAACAATAGAAATAATGGCGTGGTAACAGATGTCACGTCGGGCACGAGTACGCCGACGCTGTCCCCGCGCAAGGAGTTGAATGGCCGAGGGGGTACCACATCAGGGGCCTGGTCAGAGGGCGAGGGATCGTCCGCCGGTGCCTCAGTGCACAGCGACGAGGACGACACACAGGCTATATTGGAGTTAATGTCTACTGCTCGTAAACCACAGGTCAGCGACATGACACCTACCAGTGCCTTAGTGCAGTGGAACTCTCAACTTCCGGAAGGTGTCCCGCCGCCCAACCTTGAATTAACCTACGATCTAATACTCGGAGATCGTGGCCGGTACAAAGCCATCTACAGCGGCCCCTCTCTTTCTTGCAGAGTTCGTGATCTGCGGCCGGGATGCGAGTACTCGGTGTACCTGCAGATCCGCCCCGGGGCCGTCACGGGCGCCACCACGGAGGCGGTGACGTTCCGCGCGCCCGCCGCCCCGCCCGACCGCGTGCCCGCCCCCAAGGTCACGCAGCGGTCGCGCAACTCGCTGCTGTTGCGCTGGCCCTCCGCCGCCGACAACGGAGCGCGTCTGCAGTACTACGTGCTCGAAATGGACGACGGCGACGGCTTCAAGGAGATATCGCGCCCGCGCACTCGCCAGCACTCCGCTACCGGCCTGCGTCCCCAAACGCGATACATCTTCCGCATCGCCGCTGTTAACGAGTGCGGTCGAGGTGAATGGAGTGAAGAGATAACCGTGTGGACTTACGGGTTGCCACCCCCCGCGCCCGCTCCGCCCGTGCTCGACTCTGCCGCCTCCGAATCTCTATCTCTGTCTTGGGAACGCCGAGGCGAAGAAGAATTCACGCTCCAAATGGACGATATCGCTCGCGGGCACGGTTTCCTGCCCGTCTACAATGGTCCAGATCGAACGCACGTGTGTAAGGGTCTGCGTCGTGCATCCGACTACCGTTTTAGATTATGCTGCGAGACCACCGACGGACAAGGTCCTTATTCGACAGATGTGACGTACCGCACCCTACCCGAACGCCCTGGCGCACCAGGTCGACCCTCTGCCAAGGGCAAGATACATTCCCGAGCATTTCGCTTGCGGTGGGAGCCGCCTTCCGATGACGGAGGAGCCCCGGTAGAACAATACACTCTCGAACTCGATGCTGGCGATGGATTCCAGTCCGCGTACTCCGGTCCTGATAGAGAAGCGCATTGTGATCGACTTCAACCAGGTACTGACTACCGCGCACGAGTGCGGTGCTTGAATTCGGCCGGCGAGAGCGACTGGTCAAGTACCGAGACTGTCACCACGGATGCCACCATTCCTGCTGCTTGCGATGCGCCCGAGCCCACGAGTGAGCCGCGCGCTACTACCCTACCTTTACGTTGGAAACCTCCCAGCTGCACCGGCGGTACCCCTCTCACCGAGTATAGAGTTGAGGTGGCAAACGCGCAAGGAATCGCCCGCCTGGCTCACGCTGGCTTAGAAACGGAATGCATAGTACGAGAATTGTCTCCCGGTCGCCCGTACCGCGTGTGGGTAACCGCGTGCAATCGGGTCGGCTCCGGCCCGCCGTCCTCCGTCCTCGAATTCGAGACTGCGGCCGCCCCCCCGGATGCTCCCGACGCGCCTATCGCGAACGTCCAAAGCCCCAACGCAGTCCAACTCGAGTGGACTGCGCCCCCCGATAACGGCGCTAATATTATAGATTATAGAGTGGAAATGAGCGCAGCCAATGTCGACGATGCCTACACAGAGATATATCGGGGGTCGGAAACGAACTGTTCCATCGATCGTCTGACACCATTCACGCCCTACTTTTTCCGAGTGTGCGCAACGAATGTCGCCGGCCGGGGAGCGTGGTCCGCGGTACGGGACGTGCTGACTCCCCGCGCACCTCCCGCAGCCCCCTCTGCTCTGCGGTATGAAGCTACTGCAGACTCACTGCGTTTGCAGTGGCGCGCGCCGGCCTGCCACGGAGCTGATATCTTACAATATCGTGTCGAGGTTGGCGACGCAGCATTCGACACTGAAGGACCCACACCCGAACGGCTCGTCGAGGGACTCGAACCAGACGTGTCGTACCGTGTTCGCGTCGCCGCCCTAAACGAACTCGGGCTCGGCGATTGGAGTGAAGAATCGCGAGCCCGCACCCGACCTCCACCACCTGTGCCACCGATACTCCGGTGCTCCCCACCCGCTCATAACCACCTCCGACTCGAGTGGGACGCAACCGGAACCGAAGGAGTACATTATTGCGTCGAGATGCGTGCCCCGGACGGTCGCGAATTTCGTCCCGTGTATCGGGGAAGTGCACGTTCCTGTAAAGTGAAAAAATTGCGAGAGTCCTGTGCCTATGCCTTCCGGATACGAGCGGCGGACGAGAGAGGCGGGCGCGGGCCGTGGTCGGAGCCGGTCACGTTCGAGACGGGCAGCGCCCCGCCCCCCGCGCCCCGCGCCCCCGTGCTGCAGCTCCTCGCGCCGCGCCTTGCCCTCGTACAGTGGGACGTCATCGACGACGCTGAATACGTTTTGCAGTCCGCCCGCGGCAAGGATGCACTCTTCAAACAGGTGTACGCGGGTCGCGACACGCAGTTCCGGCTCGAGGAGCTGGAGGCGGGCGGCGAGTACGTGGTGCGCGTGTGTGCCGTGCGCGGCGGGCTGGCGGGCGCGTGGTCGGCGCCGGCGCGTCTACTCGTACCCGTGCCCGCAGCCGCGCCGCGCGTGCGCCGCCCCCGCCCCGCGCGCCCCCTACCCGCCCGCCACGCCGCGCTCCTGATGGCGACGGGCTTCCTAGTACTGGCAGTGCTGGTGGCCGTCCTGTTGCAGCGACTAGTGGAGCCGCGCGCCTGA
Protein
MVGVGVAEAGGGGGGVPGDGYYGEYYPPEQYYVPPDMCPHPQQHPQHAHMCTVLPEYGGLPVVTSGTMMPPLMPPVLDENMRQYLMHQHAQHPHHAPHHQPPHHQPPHHQPPPHHQPPHHYGPTNGAGGPQHFYGAGYPAHFHHVPPHHMQHSPPPPIYQKDERTQRQYSKLKQKLERKNNNRNNGVVTDVTSGTSTPTLSPRKELNGRGGTTSGAWSEGEGSSAGASVHSDEDDTQAILELMSTARKPQVSDMTPTSALVQWNSQLPEGVPPPNLELTYDLILGDRGRYKAIYSGPSLSCRVRDLRPGCEYSVYLQIRPGAVTGATTEAVTFRAPAAPPDRVPAPKVTQRSRNSLLLRWPSAADNGARLQYYVLEMDDGDGFKEISRPRTRQHSATGLRPQTRYIFRIAAVNECGRGEWSEEITVWTYGLPPPAPAPPVLDSAASESLSLSWERRGEEEFTLQMDDIARGHGFLPVYNGPDRTHVCKGLRRASDYRFRLCCETTDGQGPYSTDVTYRTLPERPGAPGRPSAKGKIHSRAFRLRWEPPSDDGGAPVEQYTLELDAGDGFQSAYSGPDREAHCDRLQPGTDYRARVRCLNSAGESDWSSTETVTTDATIPAACDAPEPTSEPRATTLPLRWKPPSCTGGTPLTEYRVEVANAQGIARLAHAGLETECIVRELSPGRPYRVWVTACNRVGSGPPSSVLEFETAAAPPDAPDAPIANVQSPNAVQLEWTAPPDNGANIIDYRVEMSAANVDDAYTEIYRGSETNCSIDRLTPFTPYFFRVCATNVAGRGAWSAVRDVLTPRAPPAAPSALRYEATADSLRLQWRAPACHGADILQYRVEVGDAAFDTEGPTPERLVEGLEPDVSYRVRVAALNELGLGDWSEESRARTRPPPPVPPILRCSPPAHNHLRLEWDATGTEGVHYCVEMRAPDGREFRPVYRGSARSCKVKKLRESCAYAFRIRAADERGGRGPWSEPVTFETGSAPPPAPRAPVLQLLAPRLALVQWDVIDDAEYVLQSARGKDALFKQVYAGRDTQFRLEELEAGGEYVVRVCAVRGGLAGAWSAPARLLVPVPAAAPRVRRPRPARPLPARHAALLMATGFLVLAVLVAVLLQRLVEPRA
Summary
Uniprot
H9J9H8
A0A212F4Q6
A0A194PLU1
A0A194R327
A0A2H1W0Y8
A0A067R8N5
+ More
A0A1W4X0M2 A0A1W4WPL4 A0A1W4X0D9 A0A1Y1N1K1 A0A1Y1N1J0 A0A1Y1N1I5 A0A1Y1N4G8 A0A1L8DLR0 A0A1B0CCU4 A0A0T6B0D7 A0A139WIG5 A0A139WI77 E0VER5 A0A2R2MJW4 A0A1S3IVA9 A0A1B6D383 N6U2E8 A0A210PXG0 A0A087UCR5 K1RFS6 A0A0P5YIU7 A0A0P5PL39 A0A0P5Z1G9 A0A0P4XR60 A0A0P4YPH7 V4A3Z1 R7T3N2 A0A0P5WXY1 A0A0P5A1S0 A0A1D2M853 A0A0P5FU66 A0A0P5M575 A0A0P5G572 A0A0N7ZL91 A0A0P5I192 A0A0P5FLJ8 A0A0N8B1H2 A0A0P5Q0Q1 A0A0P5QRI9 A0A0P5GTP2 A0A0P5JHS1 A0A0P5PPT0 A0A0P5H1K3 A0A0P6FT30 A0A0P5PLN4 A0A0P5HRW4 A0A0P5HSX2 A0A0P5GJU4 A0A0P5NDX8 A0A0P5NE17 A0A0P5IG14 A0A0P5GT34 A0A0P5NAH4 A0A0P6D869 A0A0P5GBJ2 A0A0N8BA53 A0A0P5NCL2 A0A0P5N9A3 A0A0P6D1P9 A0A0N8BGM1 A0A0P5HUF1 A0A0N8BM64 A0A0N8AY73 A0A0P5M310 A0A0P6CYF8 A0A0P5NLK2 A0A0P5NGS4 A0A0P5K535 A0A0P5GB89 A0A0P5LFV4 A0A0N8BLP5 A0A0P5L0W2 A0A0P6D4T3 A0A0P5QY54 A0A0P5K0V4 A0A0P5QUW1 A0A0P5S2K9 A0A0P5RGE5 A0A0P5HB94 A0A0P6D1Q7 A0A0P6DQ92 A0A0P5QAJ5 A0A0P5HWT7 A0A0P5HN43 A0A0P5MSR8 A0A0P5M6J7 A0A0P5KI23 A0A0N8EGB5 A0A0P5L944 A0A0P5RYV0 A0A0P5KRV5
A0A1W4X0M2 A0A1W4WPL4 A0A1W4X0D9 A0A1Y1N1K1 A0A1Y1N1J0 A0A1Y1N1I5 A0A1Y1N4G8 A0A1L8DLR0 A0A1B0CCU4 A0A0T6B0D7 A0A139WIG5 A0A139WI77 E0VER5 A0A2R2MJW4 A0A1S3IVA9 A0A1B6D383 N6U2E8 A0A210PXG0 A0A087UCR5 K1RFS6 A0A0P5YIU7 A0A0P5PL39 A0A0P5Z1G9 A0A0P4XR60 A0A0P4YPH7 V4A3Z1 R7T3N2 A0A0P5WXY1 A0A0P5A1S0 A0A1D2M853 A0A0P5FU66 A0A0P5M575 A0A0P5G572 A0A0N7ZL91 A0A0P5I192 A0A0P5FLJ8 A0A0N8B1H2 A0A0P5Q0Q1 A0A0P5QRI9 A0A0P5GTP2 A0A0P5JHS1 A0A0P5PPT0 A0A0P5H1K3 A0A0P6FT30 A0A0P5PLN4 A0A0P5HRW4 A0A0P5HSX2 A0A0P5GJU4 A0A0P5NDX8 A0A0P5NE17 A0A0P5IG14 A0A0P5GT34 A0A0P5NAH4 A0A0P6D869 A0A0P5GBJ2 A0A0N8BA53 A0A0P5NCL2 A0A0P5N9A3 A0A0P6D1P9 A0A0N8BGM1 A0A0P5HUF1 A0A0N8BM64 A0A0N8AY73 A0A0P5M310 A0A0P6CYF8 A0A0P5NLK2 A0A0P5NGS4 A0A0P5K535 A0A0P5GB89 A0A0P5LFV4 A0A0N8BLP5 A0A0P5L0W2 A0A0P6D4T3 A0A0P5QY54 A0A0P5K0V4 A0A0P5QUW1 A0A0P5S2K9 A0A0P5RGE5 A0A0P5HB94 A0A0P6D1Q7 A0A0P6DQ92 A0A0P5QAJ5 A0A0P5HWT7 A0A0P5HN43 A0A0P5MSR8 A0A0P5M6J7 A0A0P5KI23 A0A0N8EGB5 A0A0P5L944 A0A0P5RYV0 A0A0P5KRV5
Pubmed
EMBL
BABH01004844
AGBW02010325
OWR48716.1
KQ459601
KPI93958.1
KQ460855
+ More
KPJ11924.1 ODYU01005662 SOQ46749.1 KK852863 KDR14794.1 GEZM01015297 JAV91772.1 GEZM01015298 JAV91771.1 GEZM01015296 JAV91773.1 GEZM01015299 JAV91770.1 GFDF01006804 JAV07280.1 AJWK01007111 AJWK01007112 LJIG01022450 KRT80542.1 KQ971342 KYB27709.1 KYB27708.1 DS235093 EEB11871.1 GEDC01017129 JAS20169.1 APGK01052459 APGK01052460 KB741212 ENN72732.1 NEDP02005419 OWF41154.1 KK119237 KFM75154.1 JH816951 EKC40245.1 GDIP01058680 JAM45035.1 GDIQ01126866 JAL24860.1 GDIP01177860 GDIP01050729 LRGB01001115 JAM52986.1 KZS13524.1 GDIP01237814 JAI85587.1 GDIP01226506 JAI96895.1 KB202793 ESO87946.1 AMQN01015903 KB312336 ELT87402.1 GDIP01080724 JAM22991.1 GDIP01205315 JAJ18087.1 LJIJ01002841 ODM89167.1 GDIQ01253628 GDIQ01253627 JAJ98096.1 GDIQ01169154 JAK82571.1 GDIQ01253625 JAJ98099.1 GDIP01234409 JAI88992.1 GDIQ01222085 GDIQ01222084 JAK29640.1 GDIQ01253626 JAJ98098.1 GDIQ01222083 JAK29642.1 GDIQ01224438 GDIQ01130861 JAL20865.1 GDIQ01130860 JAL20866.1 GDIQ01236621 JAK15104.1 GDIQ01224437 JAK27288.1 GDIQ01125166 JAL26560.1 GDIQ01236622 GDIQ01233073 GDIQ01125165 JAK15103.1 GDIQ01224439 GDIQ01200688 GDIQ01043443 JAN51294.1 GDIQ01132166 GDIQ01132165 JAL19561.1 GDIQ01223177 JAK28548.1 GDIQ01223176 JAK28549.1 GDIQ01243481 GDIQ01243480 GDIQ01243479 GDIQ01242180 GDIQ01242177 GDIQ01240775 GDIQ01240774 GDIQ01240772 GDIQ01239072 GDIQ01239027 JAK08246.1 GDIQ01146712 JAL05014.1 GDIQ01266206 GDIQ01237726 GDIQ01237725 GDIQ01146711 JAL05015.1 GDIQ01213931 GDIQ01213878 JAK37847.1 GDIQ01243477 GDIQ01242179 GDIQ01240773 GDIQ01239073 JAK08248.1 GDIQ01155848 JAK95877.1 GDIQ01199383 GDIQ01082775 JAN11962.1 GDIQ01243478 GDIQ01242178 JAK08247.1 GDIQ01197835 JAK53890.1 GDIQ01244722 GDIQ01145456 GDIQ01145454 GDIQ01144055 GDIQ01126865 GDIQ01126864 JAL07671.1 GDIQ01145455 JAL06271.1 GDIQ01084224 JAN10513.1 GDIQ01179691 JAK72034.1 GDIQ01223178 JAK28547.1 GDIQ01164147 JAK87578.1 GDIQ01231276 GDIQ01231275 JAK20450.1 GDIQ01164148 JAK87577.1 GDIQ01085779 JAN08958.1 GDIQ01140609 JAL11117.1 GDIQ01142605 GDIQ01140608 JAL09121.1 GDIQ01218035 GDIQ01217131 GDIQ01189138 GDIQ01189137 GDIQ01189136 GDIQ01187744 GDIQ01187743 GDIQ01184449 GDIQ01184448 GDIQ01157995 JAK62587.1 GDIQ01250873 JAK00852.1 GDIQ01174488 JAK77237.1 GDIQ01165499 GDIQ01165498 GDIQ01165497 GDIQ01159796 JAK86226.1 GDIQ01219176 GDIQ01206631 GDIQ01203537 GDIQ01201938 GDIQ01201937 GDIQ01201936 GDIQ01138511 GDIQ01117217 GDIQ01117216 GDIQ01117215 GDIQ01115966 GDIQ01115965 JAK49789.1 GDIQ01082776 JAN11961.1 GDIQ01128309 JAL23417.1 GDIQ01196340 JAK55385.1 GDIQ01129561 JAL22165.1 GDIQ01225632 GDIQ01099680 JAL52046.1 GDIQ01207983 GDIQ01101023 GDIQ01101022 GDIQ01101021 JAL50703.1 GDIQ01235471 GDIQ01197834 GDIQ01081225 JAK16254.1 GDIQ01084225 JAN10512.1 GDIQ01084226 JAN10511.1 GDIQ01138512 JAL13214.1 GDIQ01244725 GDIQ01244724 GDIQ01244723 JAK07000.1 GDIQ01225631 JAK26094.1 GDIQ01174489 JAK77236.1 GDIQ01162804 JAK88921.1 GDIQ01189135 GDIQ01187742 GDIQ01110465 JAK62590.1 GDIQ01029548 JAN65189.1 GDIQ01178237 GDIQ01176638 GDIQ01174490 GDIQ01062942 GDIQ01061087 JAK73488.1 GDIQ01094522 JAL57204.1 GDIQ01181087 JAK70638.1
KPJ11924.1 ODYU01005662 SOQ46749.1 KK852863 KDR14794.1 GEZM01015297 JAV91772.1 GEZM01015298 JAV91771.1 GEZM01015296 JAV91773.1 GEZM01015299 JAV91770.1 GFDF01006804 JAV07280.1 AJWK01007111 AJWK01007112 LJIG01022450 KRT80542.1 KQ971342 KYB27709.1 KYB27708.1 DS235093 EEB11871.1 GEDC01017129 JAS20169.1 APGK01052459 APGK01052460 KB741212 ENN72732.1 NEDP02005419 OWF41154.1 KK119237 KFM75154.1 JH816951 EKC40245.1 GDIP01058680 JAM45035.1 GDIQ01126866 JAL24860.1 GDIP01177860 GDIP01050729 LRGB01001115 JAM52986.1 KZS13524.1 GDIP01237814 JAI85587.1 GDIP01226506 JAI96895.1 KB202793 ESO87946.1 AMQN01015903 KB312336 ELT87402.1 GDIP01080724 JAM22991.1 GDIP01205315 JAJ18087.1 LJIJ01002841 ODM89167.1 GDIQ01253628 GDIQ01253627 JAJ98096.1 GDIQ01169154 JAK82571.1 GDIQ01253625 JAJ98099.1 GDIP01234409 JAI88992.1 GDIQ01222085 GDIQ01222084 JAK29640.1 GDIQ01253626 JAJ98098.1 GDIQ01222083 JAK29642.1 GDIQ01224438 GDIQ01130861 JAL20865.1 GDIQ01130860 JAL20866.1 GDIQ01236621 JAK15104.1 GDIQ01224437 JAK27288.1 GDIQ01125166 JAL26560.1 GDIQ01236622 GDIQ01233073 GDIQ01125165 JAK15103.1 GDIQ01224439 GDIQ01200688 GDIQ01043443 JAN51294.1 GDIQ01132166 GDIQ01132165 JAL19561.1 GDIQ01223177 JAK28548.1 GDIQ01223176 JAK28549.1 GDIQ01243481 GDIQ01243480 GDIQ01243479 GDIQ01242180 GDIQ01242177 GDIQ01240775 GDIQ01240774 GDIQ01240772 GDIQ01239072 GDIQ01239027 JAK08246.1 GDIQ01146712 JAL05014.1 GDIQ01266206 GDIQ01237726 GDIQ01237725 GDIQ01146711 JAL05015.1 GDIQ01213931 GDIQ01213878 JAK37847.1 GDIQ01243477 GDIQ01242179 GDIQ01240773 GDIQ01239073 JAK08248.1 GDIQ01155848 JAK95877.1 GDIQ01199383 GDIQ01082775 JAN11962.1 GDIQ01243478 GDIQ01242178 JAK08247.1 GDIQ01197835 JAK53890.1 GDIQ01244722 GDIQ01145456 GDIQ01145454 GDIQ01144055 GDIQ01126865 GDIQ01126864 JAL07671.1 GDIQ01145455 JAL06271.1 GDIQ01084224 JAN10513.1 GDIQ01179691 JAK72034.1 GDIQ01223178 JAK28547.1 GDIQ01164147 JAK87578.1 GDIQ01231276 GDIQ01231275 JAK20450.1 GDIQ01164148 JAK87577.1 GDIQ01085779 JAN08958.1 GDIQ01140609 JAL11117.1 GDIQ01142605 GDIQ01140608 JAL09121.1 GDIQ01218035 GDIQ01217131 GDIQ01189138 GDIQ01189137 GDIQ01189136 GDIQ01187744 GDIQ01187743 GDIQ01184449 GDIQ01184448 GDIQ01157995 JAK62587.1 GDIQ01250873 JAK00852.1 GDIQ01174488 JAK77237.1 GDIQ01165499 GDIQ01165498 GDIQ01165497 GDIQ01159796 JAK86226.1 GDIQ01219176 GDIQ01206631 GDIQ01203537 GDIQ01201938 GDIQ01201937 GDIQ01201936 GDIQ01138511 GDIQ01117217 GDIQ01117216 GDIQ01117215 GDIQ01115966 GDIQ01115965 JAK49789.1 GDIQ01082776 JAN11961.1 GDIQ01128309 JAL23417.1 GDIQ01196340 JAK55385.1 GDIQ01129561 JAL22165.1 GDIQ01225632 GDIQ01099680 JAL52046.1 GDIQ01207983 GDIQ01101023 GDIQ01101022 GDIQ01101021 JAL50703.1 GDIQ01235471 GDIQ01197834 GDIQ01081225 JAK16254.1 GDIQ01084225 JAN10512.1 GDIQ01084226 JAN10511.1 GDIQ01138512 JAL13214.1 GDIQ01244725 GDIQ01244724 GDIQ01244723 JAK07000.1 GDIQ01225631 JAK26094.1 GDIQ01174489 JAK77236.1 GDIQ01162804 JAK88921.1 GDIQ01189135 GDIQ01187742 GDIQ01110465 JAK62590.1 GDIQ01029548 JAN65189.1 GDIQ01178237 GDIQ01176638 GDIQ01174490 GDIQ01062942 GDIQ01061087 JAK73488.1 GDIQ01094522 JAL57204.1 GDIQ01181087 JAK70638.1
Proteomes
PRIDE
Pfam
PF00041 fn3
SUPFAM
SSF49265
SSF49265
Gene 3D
CDD
ProteinModelPortal
H9J9H8
A0A212F4Q6
A0A194PLU1
A0A194R327
A0A2H1W0Y8
A0A067R8N5
+ More
A0A1W4X0M2 A0A1W4WPL4 A0A1W4X0D9 A0A1Y1N1K1 A0A1Y1N1J0 A0A1Y1N1I5 A0A1Y1N4G8 A0A1L8DLR0 A0A1B0CCU4 A0A0T6B0D7 A0A139WIG5 A0A139WI77 E0VER5 A0A2R2MJW4 A0A1S3IVA9 A0A1B6D383 N6U2E8 A0A210PXG0 A0A087UCR5 K1RFS6 A0A0P5YIU7 A0A0P5PL39 A0A0P5Z1G9 A0A0P4XR60 A0A0P4YPH7 V4A3Z1 R7T3N2 A0A0P5WXY1 A0A0P5A1S0 A0A1D2M853 A0A0P5FU66 A0A0P5M575 A0A0P5G572 A0A0N7ZL91 A0A0P5I192 A0A0P5FLJ8 A0A0N8B1H2 A0A0P5Q0Q1 A0A0P5QRI9 A0A0P5GTP2 A0A0P5JHS1 A0A0P5PPT0 A0A0P5H1K3 A0A0P6FT30 A0A0P5PLN4 A0A0P5HRW4 A0A0P5HSX2 A0A0P5GJU4 A0A0P5NDX8 A0A0P5NE17 A0A0P5IG14 A0A0P5GT34 A0A0P5NAH4 A0A0P6D869 A0A0P5GBJ2 A0A0N8BA53 A0A0P5NCL2 A0A0P5N9A3 A0A0P6D1P9 A0A0N8BGM1 A0A0P5HUF1 A0A0N8BM64 A0A0N8AY73 A0A0P5M310 A0A0P6CYF8 A0A0P5NLK2 A0A0P5NGS4 A0A0P5K535 A0A0P5GB89 A0A0P5LFV4 A0A0N8BLP5 A0A0P5L0W2 A0A0P6D4T3 A0A0P5QY54 A0A0P5K0V4 A0A0P5QUW1 A0A0P5S2K9 A0A0P5RGE5 A0A0P5HB94 A0A0P6D1Q7 A0A0P6DQ92 A0A0P5QAJ5 A0A0P5HWT7 A0A0P5HN43 A0A0P5MSR8 A0A0P5M6J7 A0A0P5KI23 A0A0N8EGB5 A0A0P5L944 A0A0P5RYV0 A0A0P5KRV5
A0A1W4X0M2 A0A1W4WPL4 A0A1W4X0D9 A0A1Y1N1K1 A0A1Y1N1J0 A0A1Y1N1I5 A0A1Y1N4G8 A0A1L8DLR0 A0A1B0CCU4 A0A0T6B0D7 A0A139WIG5 A0A139WI77 E0VER5 A0A2R2MJW4 A0A1S3IVA9 A0A1B6D383 N6U2E8 A0A210PXG0 A0A087UCR5 K1RFS6 A0A0P5YIU7 A0A0P5PL39 A0A0P5Z1G9 A0A0P4XR60 A0A0P4YPH7 V4A3Z1 R7T3N2 A0A0P5WXY1 A0A0P5A1S0 A0A1D2M853 A0A0P5FU66 A0A0P5M575 A0A0P5G572 A0A0N7ZL91 A0A0P5I192 A0A0P5FLJ8 A0A0N8B1H2 A0A0P5Q0Q1 A0A0P5QRI9 A0A0P5GTP2 A0A0P5JHS1 A0A0P5PPT0 A0A0P5H1K3 A0A0P6FT30 A0A0P5PLN4 A0A0P5HRW4 A0A0P5HSX2 A0A0P5GJU4 A0A0P5NDX8 A0A0P5NE17 A0A0P5IG14 A0A0P5GT34 A0A0P5NAH4 A0A0P6D869 A0A0P5GBJ2 A0A0N8BA53 A0A0P5NCL2 A0A0P5N9A3 A0A0P6D1P9 A0A0N8BGM1 A0A0P5HUF1 A0A0N8BM64 A0A0N8AY73 A0A0P5M310 A0A0P6CYF8 A0A0P5NLK2 A0A0P5NGS4 A0A0P5K535 A0A0P5GB89 A0A0P5LFV4 A0A0N8BLP5 A0A0P5L0W2 A0A0P6D4T3 A0A0P5QY54 A0A0P5K0V4 A0A0P5QUW1 A0A0P5S2K9 A0A0P5RGE5 A0A0P5HB94 A0A0P6D1Q7 A0A0P6DQ92 A0A0P5QAJ5 A0A0P5HWT7 A0A0P5HN43 A0A0P5MSR8 A0A0P5M6J7 A0A0P5KI23 A0A0N8EGB5 A0A0P5L944 A0A0P5RYV0 A0A0P5KRV5
PDB
2CRM
E-value=1.85455e-20,
Score=249
Ontologies
GO
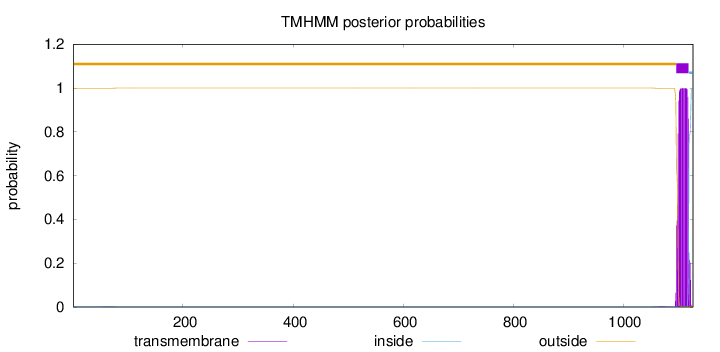
Topology
Length:
1127
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.67113
Exp number, first 60 AAs:
0.00666
Total prob of N-in:
0.00084
outside
1 - 1096
TMhelix
1097 - 1119
inside
1120 - 1127
Population Genetic Test Statistics
Pi
196.904469
Theta
165.949003
Tajima's D
0.665947
CLR
0.509222
CSRT
0.564921753912304
Interpretation
Uncertain
