Gene
KWMTBOMO01811
Pre Gene Modal
BGIBMGA005988
Annotation
sodium/solute_symporter_[Danaus_plexippus]
Full name
Cation-transporting ATPase
Location in the cell
Nuclear Reliability : 3.35
Sequence
CDS
ATGGTCTTCACGATGAAAGAAGGAGGTTTTGGGGTGGGCGATTATGTGGCGTTTGGAGTGCTATGTTCAGCGTCATGTGCTGGAGGCATTTGGTACAGTGCCGTTGGATCTAGAACTAAAGCTGTCGTAGACATCAAGGATTACCTCCTGGGAGGAAGAGCCATGTCTACTTTCCCAGTTGCTATGTCGCTTATAGCAAGTTACGTGTCAGGGGTGACGATCCTGGGTACGCCCGCGGAGATCTACAACTACGGGACGCAGTACTGGTTGGTGATCGTCGGTGTCACACTCAGCTGTGTGGTGGTAGCTACAGTTTACTTGCCCGTATTCTGTACGTTACGGCTTTCGTCTTCGTATGAATATCTAGAATTGAGATTCAATCGCAACGTGCGTGCCGTCGCTTCTATTCTATTTTTATTGGACGAGGTGTTGTTTCTACCGATGGTTGTTTATGTTCCGGCATTAGCTTTTAATCAATTGACAGGATTCAATGTCTTTGCCGTAGGTGGTATTATGGTTGTCATATGTGGGCTTTACACAGTATTAGGTGGTCTCCGCGCCGTGGTATGGACGGATTCAGTACAGACCGGTGTTATGTTCATCGGAGTGTTCCTGGTGGCGGCCGCCGGCACCATCGCTGTGGGCGGAGTGAAGGCGATCATCGATGTCGCCAATCAATCAGGGAGATTGCAATTGTCAAATTGGAACTTTTCGCCATACGAACGGCAGACCGGCTGGGGCGCTATATTCGGCGGCTTCCTCTACTGGACCTGTTTCAACTCCGTCAATCAGACCATGGTTCAGCGTTACATAGCCCTCTCTACCAAAAGGAAAGCTATTACCGCACTAGGAATATTCTGTATCGGTGCGATACTGGCTATATCGCTGTGCGTGTGGTGCGGTCTGGCCGCGTGGACAGCGTGGGTGACAGGAGGGTGCAGTCCCGGCGGAGCACCGCTAGTGGACGATAGGTTACTGCCGGCTTTTGTTACATACGTGGCGAGGATGCAGCATTTACCTGGACTGGCAGGTGTTTTCTTGGCCGGAGTGTTTGGAGCCGGATTGAGTTCGCTGTCAGCAGTTCTCAATGCATGTGCCCTAGTGGCAGTGGAAGACATCCTTAGGGGTTGGTTGCGCATAAGGCTCCGGCCGCTCGTCGAGGGGCTAATCGCGAGACTGTGCACCGGAATACTGGCGATCGTTTCCGTGGTCATGTTGATCGTCATTGAGAAACTTGGAGGCGTGCTTGGTGTGGCAACGGCTCTATCAGCAATCGCAGCGAGCACAACATGCGGCATATTCACGCTGGGCATGGTCTGCTGGTGGGTGGGACCTCGAGGAGCCATAGCGGGAGGGATAGCCGGGGCCCTAGTGGCCGGCACTACGTCGTTAGGAACTCAGGCGGCTGCCGCCAAAGGACTGAGGTCGCCGCCTCTGAACATCACAGCCGAATGCGCTAGAAACGCGACTGTCGTTTCTGTCTCTGATTTGGATCCAGAGACAATATTCCCTCTATTCCGGGTATCGTACCACTGGATAGCGCCGCTGGGCCTCGTCAGCACGCTTATAGTGGGCGCCCTGGTCGGTTGGTTGTTCGATAAGAAACAGACGAACAAGATGGACGCGGAACTATTCACTCCGGTGATATGGAGGTGGCTACCGCGCGAGGCGCACGACAACGCTGGCGTGACGCGGCGCACGTTGTCGCCGATCGACGAGTCGCCGGCGCCCTCCTCGCCGCTAATACTCGCCAAACTCGACAAGATCGAAGTTAACGGAGAGACAAGTAAATGA
Protein
MVFTMKEGGFGVGDYVAFGVLCSASCAGGIWYSAVGSRTKAVVDIKDYLLGGRAMSTFPVAMSLIASYVSGVTILGTPAEIYNYGTQYWLVIVGVTLSCVVVATVYLPVFCTLRLSSSYEYLELRFNRNVRAVASILFLLDEVLFLPMVVYVPALAFNQLTGFNVFAVGGIMVVICGLYTVLGGLRAVVWTDSVQTGVMFIGVFLVAAAGTIAVGGVKAIIDVANQSGRLQLSNWNFSPYERQTGWGAIFGGFLYWTCFNSVNQTMVQRYIALSTKRKAITALGIFCIGAILAISLCVWCGLAAWTAWVTGGCSPGGAPLVDDRLLPAFVTYVARMQHLPGLAGVFLAGVFGAGLSSLSAVLNACALVAVEDILRGWLRIRLRPLVEGLIARLCTGILAIVSVVMLIVIEKLGGVLGVATALSAIAASTTCGIFTLGMVCWWVGPRGAIAGGIAGALVAGTTSLGTQAAAAKGLRSPPLNITAECARNATVVSVSDLDPETIFPLFRVSYHWIAPLGLVSTLIVGALVGWLFDKKQTNKMDAELFTPVIWRWLPREAHDNAGVTRRTLSPIDESPAPSSPLILAKLDKIEVNGETSK
Summary
Similarity
Belongs to the sodium:solute symporter (SSF) (TC 2.A.21) family.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type V subfamily.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type V subfamily.
Uniprot
H9J8Z6
A0A194PKX1
A0A2A4JHE6
A0A2H1VBX4
A0A182GSX3
Q17LX4
+ More
A0A1Q3FBN6 A0A1Q3FET1 A0A1Q3FEW4 T1P9Q6 B0XK25 W5JBC7 A0A2M4A618 Q7PIZ6 A0A2M4A5Z1 A0A2M3ZKP7 A0A2C9H8R6 A0A1S4GWD1 A0A2Y9D152 W8ARK5 A0A336LK96 A0A2M4BGL2 A0A2M4BGJ1 A0A182J9A1 A0A0A1WPR9 A0A084VY64 A0A182L2F5 A0A182F115 A0A182QQW1 A0A182MZF9 A0A0L0CBS1 A0A1Y1N0Q0 B4LUB9 A0A0Q9WKN6 A0A1I8Q542 A0A1I8Q545 A0A0M3QT38 A0A182R3B6 E2A0B6 B4KGW6 A0A182TME0 A0A182VA30 A0A0K8VS11 A0A0Q9X3F9 A0A182P0H2 A0A0K8V9L1 A0A034VEH2 A0A195FKU8 A0A1B0A5W9 A0A034VDC7 A0A1S4GWD6 A0A182YGS0 A0A151WSM8 A0A182K6Z5 A0A182LWM8 A0A1B0FEN1 B4JB31 A0A1A9VHU9 A0A195BW95 A0A195CCU2 A0A151J0D7 A0A1L8DDW0 F4WJB8 A0A2C9H8Q8 B3MPI4 A0A1S4GX53 A0A1W4UUW4 A0A1W4V8J7 B4MVX3 B4NWF0 A0A1A9X648 A0A0A1WWL8 A0A0J9QZ62 Q9VLR8 B3N717 Q29ML4 A0A310S8Y6 A0A1S4GWC9 B4HYG1 D6WKE2 A0A2C9H8R5 A0A3B0K2E1 A0A1J1IPE0 A0A158NEA5 A0A139WIB2 K7IYN9 A0A1B0CYG7 A0A2Y9D116 A0A3L8DC01 A0A0L7QQ94 A0A026W499 A0A2Y9D166 B4PU64 A0A023EUD0 A0A182WA35 B3P3F9 A0A232F2E3
A0A1Q3FBN6 A0A1Q3FET1 A0A1Q3FEW4 T1P9Q6 B0XK25 W5JBC7 A0A2M4A618 Q7PIZ6 A0A2M4A5Z1 A0A2M3ZKP7 A0A2C9H8R6 A0A1S4GWD1 A0A2Y9D152 W8ARK5 A0A336LK96 A0A2M4BGL2 A0A2M4BGJ1 A0A182J9A1 A0A0A1WPR9 A0A084VY64 A0A182L2F5 A0A182F115 A0A182QQW1 A0A182MZF9 A0A0L0CBS1 A0A1Y1N0Q0 B4LUB9 A0A0Q9WKN6 A0A1I8Q542 A0A1I8Q545 A0A0M3QT38 A0A182R3B6 E2A0B6 B4KGW6 A0A182TME0 A0A182VA30 A0A0K8VS11 A0A0Q9X3F9 A0A182P0H2 A0A0K8V9L1 A0A034VEH2 A0A195FKU8 A0A1B0A5W9 A0A034VDC7 A0A1S4GWD6 A0A182YGS0 A0A151WSM8 A0A182K6Z5 A0A182LWM8 A0A1B0FEN1 B4JB31 A0A1A9VHU9 A0A195BW95 A0A195CCU2 A0A151J0D7 A0A1L8DDW0 F4WJB8 A0A2C9H8Q8 B3MPI4 A0A1S4GX53 A0A1W4UUW4 A0A1W4V8J7 B4MVX3 B4NWF0 A0A1A9X648 A0A0A1WWL8 A0A0J9QZ62 Q9VLR8 B3N717 Q29ML4 A0A310S8Y6 A0A1S4GWC9 B4HYG1 D6WKE2 A0A2C9H8R5 A0A3B0K2E1 A0A1J1IPE0 A0A158NEA5 A0A139WIB2 K7IYN9 A0A1B0CYG7 A0A2Y9D116 A0A3L8DC01 A0A0L7QQ94 A0A026W499 A0A2Y9D166 B4PU64 A0A023EUD0 A0A182WA35 B3P3F9 A0A232F2E3
EC Number
3.6.3.-
Pubmed
19121390
26354079
26483478
17519023
17510324
25315136
+ More
20920257 23761445 12364791 24495485 25830018 24438588 20966253 26108605 28004739 17994087 20798317 25348373 25244985 21719571 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 21347285 20075255 30249741 24508170 24945155 28648823
20920257 23761445 12364791 24495485 25830018 24438588 20966253 26108605 28004739 17994087 20798317 25348373 25244985 21719571 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 21347285 20075255 30249741 24508170 24945155 28648823
EMBL
BABH01004822
BABH01004823
BABH01004824
BABH01004825
KQ459601
KPI93977.1
+ More
NWSH01001371 PCG71495.1 ODYU01001724 SOQ38315.1 JXUM01085578 JXUM01085579 KQ563511 KXJ73721.1 EF173367 CH477210 ABM68585.1 EAT47697.1 GFDL01010050 JAV24995.1 GFDL01008986 JAV26059.1 GFDL01008955 JAV26090.1 KA645429 AFP60058.1 DS233684 EDS31122.1 ADMH02001963 ETN60160.1 GGFK01002880 MBW36201.1 AAAB01008964 EAA43957.4 GGFK01002903 MBW36224.1 GGFM01008360 MBW29111.1 APCN01005204 GAMC01017858 GAMC01017854 JAB88697.1 UFQT01000036 SSX18360.1 GGFJ01003049 MBW52190.1 GGFJ01003048 MBW52189.1 GBXI01015213 GBXI01013626 JAC99078.1 JAD00666.1 ATLV01018323 KE525231 KFB42908.1 AXCN02000606 JRES01000736 KNC28884.1 GEZM01015657 JAV91483.1 CH940649 EDW64105.1 KRF81419.1 CP012523 ALC38209.1 GL435569 EFN73119.1 CH933807 EDW11166.1 GDHF01023020 GDHF01010647 JAI29294.1 JAI41667.1 KRG02560.1 GDHF01017084 GDHF01007157 JAI35230.1 JAI45157.1 GAKP01018445 GAKP01018440 JAC40507.1 KQ981490 KYN41043.1 GAKP01018442 GAKP01018438 JAC40514.1 KQ982769 KYQ50838.1 AXCM01001018 CCAG010014746 CH916368 EDW03923.1 KQ976401 KYM92555.1 KQ978023 KYM97903.1 KQ980636 KYN14958.1 GFDF01009527 JAV04557.1 GL888182 EGI65696.1 CH902620 EDV31280.1 CH963857 EDW75843.2 CM000157 EDW88467.1 GBXI01011246 JAD03046.1 CM002910 KMY88974.1 AE014134 AY058420 AAF52616.2 AAL13649.1 CH954177 EDV59313.1 CH379060 EAL33679.3 KQ763867 OAD54694.1 CH480818 EDW52091.1 KQ971342 EFA03978.2 OUUW01000004 SPP79776.1 CVRI01000055 CRL01594.1 ADTU01013221 KYB27649.1 AJVK01000161 AJVK01000162 QOIP01000010 RLU17861.1 KQ414786 KOC60800.1 KK107455 EZA50426.1 CM000160 EDW97714.2 KRK03925.1 GAPW01001046 JAC12552.1 CH954181 EDV48739.1 NNAY01001175 OXU24865.1
NWSH01001371 PCG71495.1 ODYU01001724 SOQ38315.1 JXUM01085578 JXUM01085579 KQ563511 KXJ73721.1 EF173367 CH477210 ABM68585.1 EAT47697.1 GFDL01010050 JAV24995.1 GFDL01008986 JAV26059.1 GFDL01008955 JAV26090.1 KA645429 AFP60058.1 DS233684 EDS31122.1 ADMH02001963 ETN60160.1 GGFK01002880 MBW36201.1 AAAB01008964 EAA43957.4 GGFK01002903 MBW36224.1 GGFM01008360 MBW29111.1 APCN01005204 GAMC01017858 GAMC01017854 JAB88697.1 UFQT01000036 SSX18360.1 GGFJ01003049 MBW52190.1 GGFJ01003048 MBW52189.1 GBXI01015213 GBXI01013626 JAC99078.1 JAD00666.1 ATLV01018323 KE525231 KFB42908.1 AXCN02000606 JRES01000736 KNC28884.1 GEZM01015657 JAV91483.1 CH940649 EDW64105.1 KRF81419.1 CP012523 ALC38209.1 GL435569 EFN73119.1 CH933807 EDW11166.1 GDHF01023020 GDHF01010647 JAI29294.1 JAI41667.1 KRG02560.1 GDHF01017084 GDHF01007157 JAI35230.1 JAI45157.1 GAKP01018445 GAKP01018440 JAC40507.1 KQ981490 KYN41043.1 GAKP01018442 GAKP01018438 JAC40514.1 KQ982769 KYQ50838.1 AXCM01001018 CCAG010014746 CH916368 EDW03923.1 KQ976401 KYM92555.1 KQ978023 KYM97903.1 KQ980636 KYN14958.1 GFDF01009527 JAV04557.1 GL888182 EGI65696.1 CH902620 EDV31280.1 CH963857 EDW75843.2 CM000157 EDW88467.1 GBXI01011246 JAD03046.1 CM002910 KMY88974.1 AE014134 AY058420 AAF52616.2 AAL13649.1 CH954177 EDV59313.1 CH379060 EAL33679.3 KQ763867 OAD54694.1 CH480818 EDW52091.1 KQ971342 EFA03978.2 OUUW01000004 SPP79776.1 CVRI01000055 CRL01594.1 ADTU01013221 KYB27649.1 AJVK01000161 AJVK01000162 QOIP01000010 RLU17861.1 KQ414786 KOC60800.1 KK107455 EZA50426.1 CM000160 EDW97714.2 KRK03925.1 GAPW01001046 JAC12552.1 CH954181 EDV48739.1 NNAY01001175 OXU24865.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000069940
UP000249989
UP000008820
+ More
UP000095301 UP000002320 UP000000673 UP000007062 UP000076407 UP000075840 UP000075880 UP000030765 UP000075882 UP000069272 UP000075886 UP000075884 UP000037069 UP000008792 UP000095300 UP000092553 UP000075900 UP000000311 UP000009192 UP000075902 UP000075903 UP000075885 UP000078541 UP000092445 UP000076408 UP000075809 UP000075881 UP000075883 UP000092444 UP000001070 UP000078200 UP000078540 UP000078542 UP000078492 UP000007755 UP000007801 UP000192221 UP000007798 UP000002282 UP000092443 UP000000803 UP000008711 UP000001819 UP000001292 UP000007266 UP000268350 UP000183832 UP000005205 UP000002358 UP000092462 UP000279307 UP000053825 UP000053097 UP000075920 UP000215335
UP000095301 UP000002320 UP000000673 UP000007062 UP000076407 UP000075840 UP000075880 UP000030765 UP000075882 UP000069272 UP000075886 UP000075884 UP000037069 UP000008792 UP000095300 UP000092553 UP000075900 UP000000311 UP000009192 UP000075902 UP000075903 UP000075885 UP000078541 UP000092445 UP000076408 UP000075809 UP000075881 UP000075883 UP000092444 UP000001070 UP000078200 UP000078540 UP000078542 UP000078492 UP000007755 UP000007801 UP000192221 UP000007798 UP000002282 UP000092443 UP000000803 UP000008711 UP000001819 UP000001292 UP000007266 UP000268350 UP000183832 UP000005205 UP000002358 UP000092462 UP000279307 UP000053825 UP000053097 UP000075920 UP000215335
Pfam
PF00474 SSF
Interpro
Gene 3D
ProteinModelPortal
H9J8Z6
A0A194PKX1
A0A2A4JHE6
A0A2H1VBX4
A0A182GSX3
Q17LX4
+ More
A0A1Q3FBN6 A0A1Q3FET1 A0A1Q3FEW4 T1P9Q6 B0XK25 W5JBC7 A0A2M4A618 Q7PIZ6 A0A2M4A5Z1 A0A2M3ZKP7 A0A2C9H8R6 A0A1S4GWD1 A0A2Y9D152 W8ARK5 A0A336LK96 A0A2M4BGL2 A0A2M4BGJ1 A0A182J9A1 A0A0A1WPR9 A0A084VY64 A0A182L2F5 A0A182F115 A0A182QQW1 A0A182MZF9 A0A0L0CBS1 A0A1Y1N0Q0 B4LUB9 A0A0Q9WKN6 A0A1I8Q542 A0A1I8Q545 A0A0M3QT38 A0A182R3B6 E2A0B6 B4KGW6 A0A182TME0 A0A182VA30 A0A0K8VS11 A0A0Q9X3F9 A0A182P0H2 A0A0K8V9L1 A0A034VEH2 A0A195FKU8 A0A1B0A5W9 A0A034VDC7 A0A1S4GWD6 A0A182YGS0 A0A151WSM8 A0A182K6Z5 A0A182LWM8 A0A1B0FEN1 B4JB31 A0A1A9VHU9 A0A195BW95 A0A195CCU2 A0A151J0D7 A0A1L8DDW0 F4WJB8 A0A2C9H8Q8 B3MPI4 A0A1S4GX53 A0A1W4UUW4 A0A1W4V8J7 B4MVX3 B4NWF0 A0A1A9X648 A0A0A1WWL8 A0A0J9QZ62 Q9VLR8 B3N717 Q29ML4 A0A310S8Y6 A0A1S4GWC9 B4HYG1 D6WKE2 A0A2C9H8R5 A0A3B0K2E1 A0A1J1IPE0 A0A158NEA5 A0A139WIB2 K7IYN9 A0A1B0CYG7 A0A2Y9D116 A0A3L8DC01 A0A0L7QQ94 A0A026W499 A0A2Y9D166 B4PU64 A0A023EUD0 A0A182WA35 B3P3F9 A0A232F2E3
A0A1Q3FBN6 A0A1Q3FET1 A0A1Q3FEW4 T1P9Q6 B0XK25 W5JBC7 A0A2M4A618 Q7PIZ6 A0A2M4A5Z1 A0A2M3ZKP7 A0A2C9H8R6 A0A1S4GWD1 A0A2Y9D152 W8ARK5 A0A336LK96 A0A2M4BGL2 A0A2M4BGJ1 A0A182J9A1 A0A0A1WPR9 A0A084VY64 A0A182L2F5 A0A182F115 A0A182QQW1 A0A182MZF9 A0A0L0CBS1 A0A1Y1N0Q0 B4LUB9 A0A0Q9WKN6 A0A1I8Q542 A0A1I8Q545 A0A0M3QT38 A0A182R3B6 E2A0B6 B4KGW6 A0A182TME0 A0A182VA30 A0A0K8VS11 A0A0Q9X3F9 A0A182P0H2 A0A0K8V9L1 A0A034VEH2 A0A195FKU8 A0A1B0A5W9 A0A034VDC7 A0A1S4GWD6 A0A182YGS0 A0A151WSM8 A0A182K6Z5 A0A182LWM8 A0A1B0FEN1 B4JB31 A0A1A9VHU9 A0A195BW95 A0A195CCU2 A0A151J0D7 A0A1L8DDW0 F4WJB8 A0A2C9H8Q8 B3MPI4 A0A1S4GX53 A0A1W4UUW4 A0A1W4V8J7 B4MVX3 B4NWF0 A0A1A9X648 A0A0A1WWL8 A0A0J9QZ62 Q9VLR8 B3N717 Q29ML4 A0A310S8Y6 A0A1S4GWC9 B4HYG1 D6WKE2 A0A2C9H8R5 A0A3B0K2E1 A0A1J1IPE0 A0A158NEA5 A0A139WIB2 K7IYN9 A0A1B0CYG7 A0A2Y9D116 A0A3L8DC01 A0A0L7QQ94 A0A026W499 A0A2Y9D166 B4PU64 A0A023EUD0 A0A182WA35 B3P3F9 A0A232F2E3
PDB
5NVA
E-value=3.3153e-10,
Score=158
Ontologies
GO
Topology
Subcellular location
Membrane
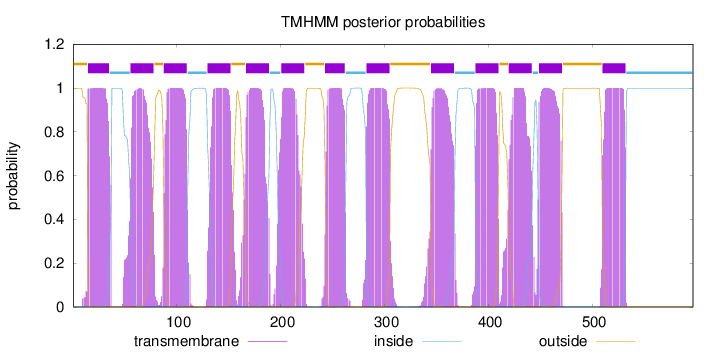
Length:
597
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
290.78416
Exp number, first 60 AAs:
28.31825
Total prob of N-in:
0.00132
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 35
inside
36 - 55
TMhelix
56 - 78
outside
79 - 87
TMhelix
88 - 110
inside
111 - 129
TMhelix
130 - 152
outside
153 - 166
TMhelix
167 - 189
inside
190 - 200
TMhelix
201 - 223
outside
224 - 242
TMhelix
243 - 262
inside
263 - 282
TMhelix
283 - 305
outside
306 - 344
TMhelix
345 - 367
inside
368 - 387
TMhelix
388 - 410
outside
411 - 419
TMhelix
420 - 442
inside
443 - 448
TMhelix
449 - 471
outside
472 - 509
TMhelix
510 - 532
inside
533 - 597
Population Genetic Test Statistics
Pi
287.38085
Theta
180.932001
Tajima's D
2.079297
CLR
0.369615
CSRT
0.895955202239888
Interpretation
Uncertain
