Gene
KWMTBOMO01806 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006162
Annotation
PREDICTED:_manganese-transporting_ATPase_13A1_[Plutella_xylostella]
Full name
Cation-transporting ATPase
Location in the cell
Nuclear Reliability : 3.738
Sequence
CDS
ATGGTAAAGATGCAGTCGTCACTAGATGATTTAGTTCAGTATACACAGCTTTTCAAGCCTGTTCCGACTGTTTTACAAGGCACGGTTTTGCCTTTTCTATTTATTTACCCTCTGGTTTTTTATTCATGGATCGTTGTGTACGGTATAGAAGATAATTTCGAGGCAGGATTCGTGACTGTGGCTGTGATAGCAACAGTCCAGATCCTCATATGTTTGTGTTGTTACTGGAGTGTTCATATCAACTGCTTCCTTTCTTGTTCAGCGGTTAAAGATCCAATACAAGCTGAGGTAGTCAAGGTTGTACCGACTTCTAACAATGGATCATCTGAGCTTGTTCGACTACATCATACGAAGTCAACAAAAAAAGACAGTATACACCCAGATGTTTGGTTCATATTTCAAAAGAGTAAATATGTGTATGACTGGGATAAAAAGAATTTTCACACCATAGAATTTCCTATCAATAAAATTTATGAAGAATATATGGAATCCAAAGGATATTCTGATGATGAGGCTATTGATTTAGCAGAAAAAGAATTTGGTAAAAATGAGATGGTTATGGTTGTTCCGGAATTTATGGAACTTTTTAAAGAGAGAGCAACAGCACCATTTTTTGTATTTCAAGTATTTTGTGTAGCTCTGTGGTGTTTGGATAAGTATTGGTATTACTCAATATTCACACTAGTGATGTTGGTTATGTTTGAATGTACATTGGTGCAGCAACAGCTCAGAAATATGGCTGAGATCAGAAAGATGGGCAATAAGCCTTACAACATTAATGTATATCGAAACAGGAGGTGGAGACAAATAACAAGCGATCAGCTTTTACCGGGTGACATAGTATCCCTCACAAGATCCGTCAACGAAAACCTGGTCCCTTGTGACATAATACTCTTACGGGGGTCCTGTATTGTAGATGAGTCCATGTTGACTGGTGAGAGTGTTCCACAAATGAAAGAAGCTTTGGAAAATGAAAAGGACCTCCAGCAACATCTAGAACCTGAGGGTGATGGTAAACTTCACATGCTTTTTGGTGGTACTAAGATTGTACAACATTCCTCACCATCCAAAAGCTCACCTTCAGGGCTTAAAGCCCCGGATAATGGTTGCATTGGTTATGTTATACGTAATGGTTTTAACACATCACAGGGTAAACTGCTAAGAACCATTTTGTTTGGAGTGAAAAGGGTCACAGCTAATAATCTTGAAACTTTTGGATTCATAATGTTTTTGCTTATATTTGCTGTAGCTGCTGCGGCCTATGTGTGGGTCAAGGGTTGCGAAGATCCTGAGCGAAATAGATACAAATTATTTTTGGAATGTACTTTAATATTGACATCGGTTGTTCCGCCTGAACTGCCCATAGAGTTGTCTCTAGCAGTTAATACGTCATTGTTGTCTTTATCGAAATTGGCCGTATTCTGTACAGAACCATTTAGGATACCCTTTGCTGGTAAAGTAGAAATATGTTGTTTCGATAAGACTGGAACATTGACAAGTGATAATTTAGTTGTTGAAGGAGTAGCTGGAATAGGAGAACACAAAGATGCAACAGTGGTTCCCCTTTCTGAAGCTCCAATGGAGACTGTTCAAGTTTTAGCATCGTGCCACTCTTTAGTTCAACTTGACGATGGAATTGTTGGGGATCCATTAGAAAAAGCTACATTGAAGGCAGCAGAGTGGAACCTCACCAAAAGCGATGCTGTAGTACCAAAGAAGGGGAAGTCACCTGGCCTGAAAATTGTTCATAGAAATCATTTTTCTAGTGCTCTCAAAAGAATGTCCGTGATAGCAGGTTATCAGATCAATGAAAGAGGTTTTATTGAGACTCACTACATCAGTAGTGTTAAGGGTGCCCCAGAAACAATCAAATCGATGCTTAAGGATGTCCCTAGTCATTATGATCATGTTTATCTAACGCTCTCGAGGAGAGGCGCGCGAGTTTTAGCTCTGGGTTATAGAAATTTAGGAAAATTGACATCTCAAGAGATAAGGGATTTATCCCGGGATGACATTGAATGTGACCTAACGTTTGTAGGTTTCGTCATAATATCATGTCCATTAAAAACGGACTCAAAGAAAGCTATAGCTGAAATTGTTCACGCTTCGCATTCAGTCGTGATGATAACGGGTGATAATCCGCTGACGGCGTGTCATGTAGCCAAAGAATTAAAATTCACCCAAAAAGAGGAGGTTTTGATATTGACCGAGAGTGATGGGGAGTGGGCATGGAAATCAATTGATGAAGAACTAAATCTTCCAGTGCAACCGTCTCAGAAAACAAAATCGTTGACCTCGAAATTCGATCTTTGCATAACCGGTGAGGGTCTGACTTTCTTGAATGAAAATCATCGTAAGTTTCTCCTCGAATTGATGCCGCACATCAAGGTTTTTGCCAGATTCGCGCCCAAGCAGAAGGAATTCGTTGTGGTCACGTTAAAGTCTTTGGGATATACCACGCTCATGTGCGGCGATGGAACTAATGATGTCGGAGCGTTAAAACATGCTGATGTCGGAGTGGCTATTTTGGCTAATGCACCAGAACGAATCCGCGAGAAACCTCGAGAGGAACGTCCTCCGGAGCCGGAGCGGGGGCGGCGCCCGAGGGACCCGCGCGCCGACGCACGAGCCGACGCCGCCGCCAGGTTCCGCCGCGCCATGAAGGAACTCGAAGACGACGACCAGCCGCAGGTCGTGAGGCTCGGAGACGCTAGCGTTGCGGCTCCCTTTACCAGCCGACTCTCAAGTATACTTTGCATTTGTCACATCATCAAACAAGGCCGTTGCACGCTGGTGACCACGCTGCAGATGTTCAAGATCCTAGCGCTGAACGCTTTGATATTGGCGTACAGTCAATCCGTGTTATATCTAGACGGAATCAAATTCAGTGACACCCAAGCGACGTTGCAGAGTTTGTTGTTGGCGTCCTGTTTCTTGTTCATTTCGAGGTCTAAGCCGTTGAAGCAGTTATCCAAAGAGCGTCCATTGCCAAATATATTCAATATGTATACAATCATGACGGTGCTCAGTCAGTTCGCGGTCCACTTCCTGTGTCTCATTTATTTGGTGCACGAGGCTACCGTGAGATCTCCGGGCCGGGACAATAAACCGAAATTAGATATGGATTTAGCTGAAGACGAAGATCGAGAATTCGCTCCAGATTTACTGAACAGCACCGTTTATATTATATCAATGGCGTTACAGATTTCCACTTTTGCTATCAACTACAGAGGTGAACCGTTCATGCAAGGTTTGAAGGACAACAAACCATTACTGTACAGCATTGTGATATCCGGAGGGGCGGTGCTTGCTCTCGCAGCCGGGATATTCCCTGACCTCTCCAACATGTTCGAAATAGTATACTTCCCACCGGATTATCGGGTGATACTCGTGCAAGTTTTGATAGCTGATATGTTGTTCGCGTATTTGGTGGACCGTTTCTGTTTGTTCCTCTTCGGGGAAGGGCGGATTCCTAAGACGTAA
Protein
MVKMQSSLDDLVQYTQLFKPVPTVLQGTVLPFLFIYPLVFYSWIVVYGIEDNFEAGFVTVAVIATVQILICLCCYWSVHINCFLSCSAVKDPIQAEVVKVVPTSNNGSSELVRLHHTKSTKKDSIHPDVWFIFQKSKYVYDWDKKNFHTIEFPINKIYEEYMESKGYSDDEAIDLAEKEFGKNEMVMVVPEFMELFKERATAPFFVFQVFCVALWCLDKYWYYSIFTLVMLVMFECTLVQQQLRNMAEIRKMGNKPYNINVYRNRRWRQITSDQLLPGDIVSLTRSVNENLVPCDIILLRGSCIVDESMLTGESVPQMKEALENEKDLQQHLEPEGDGKLHMLFGGTKIVQHSSPSKSSPSGLKAPDNGCIGYVIRNGFNTSQGKLLRTILFGVKRVTANNLETFGFIMFLLIFAVAAAAYVWVKGCEDPERNRYKLFLECTLILTSVVPPELPIELSLAVNTSLLSLSKLAVFCTEPFRIPFAGKVEICCFDKTGTLTSDNLVVEGVAGIGEHKDATVVPLSEAPMETVQVLASCHSLVQLDDGIVGDPLEKATLKAAEWNLTKSDAVVPKKGKSPGLKIVHRNHFSSALKRMSVIAGYQINERGFIETHYISSVKGAPETIKSMLKDVPSHYDHVYLTLSRRGARVLALGYRNLGKLTSQEIRDLSRDDIECDLTFVGFVIISCPLKTDSKKAIAEIVHASHSVVMITGDNPLTACHVAKELKFTQKEEVLILTESDGEWAWKSIDEELNLPVQPSQKTKSLTSKFDLCITGEGLTFLNENHRKFLLELMPHIKVFARFAPKQKEFVVVTLKSLGYTTLMCGDGTNDVGALKHADVGVAILANAPERIREKPREERPPEPERGRRPRDPRADARADAAARFRRAMKELEDDDQPQVVRLGDASVAAPFTSRLSSILCICHIIKQGRCTLVTTLQMFKILALNALILAYSQSVLYLDGIKFSDTQATLQSLLLASCFLFISRSKPLKQLSKERPLPNIFNMYTIMTVLSQFAVHFLCLIYLVHEATVRSPGRDNKPKLDMDLAEDEDREFAPDLLNSTVYIISMALQISTFAINYRGEPFMQGLKDNKPLLYSIVISGGAVLALAAGIFPDLSNMFEIVYFPPDYRVILVQVLIADMLFAYLVDRFCLFLFGEGRIPKT
Summary
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type V subfamily.
Feature
chain Cation-transporting ATPase
Uniprot
H9J9G9
A0A2H1WHX9
A0A2A4JAF7
E2JE29
A0A194PLW6
A0A212ERB9
+ More
A0A0L7LRZ6 A0A1B6DGW9 A0A194R7Q5 A0A139WHP6 A0A0N7Z9H8 V5GTV3 A0A088ALT2 A0A067QZV5 A0A2A3E3S8 A0A0M9A125 T1HIV5 A0A310SHT1 A0A151WSR6 A0A0L7QQK9 E2B656 A0A026W2W9 A0A0J7P0G2 E2A0B7 A0A195CAR3 A0A151J0E3 F4WJB7 A0A195FM34 A0A195BVA2 A0A158NEA4 A0A336KH29 Q17JE6 A0A182PMM5 A0A1B0CD08 A0A146LXI2 A0A023GMC8 A0A1J1IPE0 U5EKZ4 A0A1L8DEW8 A0A232F228 A0A1B0CYG6 T1J7Q5 A0A1L8DEQ9 E9HDF2 A0A182G5Q4 A0A1L8DE88 A0A0A9XCM8 A0A154PG96 A0A0K8SC29 A0A210PH55 E0VF24 A0A224XHB2 A0A1B0CSV1 C3ZFY8 B4KI12 A0A3B3BHL8 B4LTE4 A0A3P8SCX5 A0A3B5LYP7 B4NLP3 M4AG25 A0A0M4E649 A0A315V3K5 A0A1W5ABI2 A0A3B5AQC8 A0A0Q5WJS8 B4HX19 B4QAB2 A0A3B1KED8 B3N4L0 A0A3P9HZV1 A0A3P9JXX7 A0A1A8IBQ6 A0A3B4UT60 A0A1A8JW57 A0A1A8RNK4 A0A1A8MZQ8 A0A1A7X4V3 A0A1A8DT76 A0A3B4WE62 A0A3P9C8N9 A0A1A7ZHX3 A0A3B0JBQ6 B3MNF3 A0A3B3HYE3 A0A1A8HD03 Q9VKJ6 A0A3B4FLV9 Q29M40 A0A3B3W2A6 I3J0F7 A0A1W4W523 B4G9T9 A0A3B3X723 Q9Y139 A0A0F8B578 A0A087XMC1 B4P173 A0A0P7UU04 A0A3P9NDK9
A0A0L7LRZ6 A0A1B6DGW9 A0A194R7Q5 A0A139WHP6 A0A0N7Z9H8 V5GTV3 A0A088ALT2 A0A067QZV5 A0A2A3E3S8 A0A0M9A125 T1HIV5 A0A310SHT1 A0A151WSR6 A0A0L7QQK9 E2B656 A0A026W2W9 A0A0J7P0G2 E2A0B7 A0A195CAR3 A0A151J0E3 F4WJB7 A0A195FM34 A0A195BVA2 A0A158NEA4 A0A336KH29 Q17JE6 A0A182PMM5 A0A1B0CD08 A0A146LXI2 A0A023GMC8 A0A1J1IPE0 U5EKZ4 A0A1L8DEW8 A0A232F228 A0A1B0CYG6 T1J7Q5 A0A1L8DEQ9 E9HDF2 A0A182G5Q4 A0A1L8DE88 A0A0A9XCM8 A0A154PG96 A0A0K8SC29 A0A210PH55 E0VF24 A0A224XHB2 A0A1B0CSV1 C3ZFY8 B4KI12 A0A3B3BHL8 B4LTE4 A0A3P8SCX5 A0A3B5LYP7 B4NLP3 M4AG25 A0A0M4E649 A0A315V3K5 A0A1W5ABI2 A0A3B5AQC8 A0A0Q5WJS8 B4HX19 B4QAB2 A0A3B1KED8 B3N4L0 A0A3P9HZV1 A0A3P9JXX7 A0A1A8IBQ6 A0A3B4UT60 A0A1A8JW57 A0A1A8RNK4 A0A1A8MZQ8 A0A1A7X4V3 A0A1A8DT76 A0A3B4WE62 A0A3P9C8N9 A0A1A7ZHX3 A0A3B0JBQ6 B3MNF3 A0A3B3HYE3 A0A1A8HD03 Q9VKJ6 A0A3B4FLV9 Q29M40 A0A3B3W2A6 I3J0F7 A0A1W4W523 B4G9T9 A0A3B3X723 Q9Y139 A0A0F8B578 A0A087XMC1 B4P173 A0A0P7UU04 A0A3P9NDK9
EC Number
3.6.3.-
Pubmed
19121390
22848670
26354079
22118469
26227816
18362917
+ More
19820115 27129103 24845553 20798317 24508170 21719571 21347285 17510324 26823975 28648823 21292972 26483478 25401762 28812685 20566863 18563158 17994087 29451363 23542700 29703783 22936249 25329095 17554307 25186727 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25835551 17550304
19820115 27129103 24845553 20798317 24508170 21719571 21347285 17510324 26823975 28648823 21292972 26483478 25401762 28812685 20566863 18563158 17994087 29451363 23542700 29703783 22936249 25329095 17554307 25186727 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25835551 17550304
EMBL
BABH01004811
ODYU01008780
SOQ52680.1
NWSH01002391
PCG68383.1
HQ184468
+ More
ADN88179.1 KQ459601 KPI93983.1 AGBW02013052 OWR44040.1 JTDY01000232 KOB78149.1 GEDC01027435 GEDC01012390 JAS09863.1 JAS24908.1 KQ460855 KPJ11901.1 KQ971343 KYB27482.1 GDKW01000507 JAI56088.1 GALX01003399 JAB65067.1 KK852846 KDR15104.1 KZ288391 PBC26350.1 KQ435770 KOX75180.1 ACPB03011142 KQ763867 OAD54693.1 KQ982769 KYQ50837.1 KQ414786 KOC60801.1 GL445914 EFN88845.1 KK107455 EZA50425.1 LBMM01000533 KMQ98115.1 GL435569 EFN73120.1 KQ978023 KYM97902.1 KQ980636 KYN14959.1 GL888182 EGI65695.1 KQ981490 KYN41044.1 KQ976401 KYM92554.1 ADTU01013223 UFQS01000295 UFQT01000295 SSX02575.1 SSX22949.1 CH477233 EAT46757.1 AJWK01007292 GDHC01007247 JAQ11382.1 GBBM01000427 JAC34991.1 CVRI01000055 CRL01594.1 GANO01001647 JAB58224.1 GFDF01009210 JAV04874.1 NNAY01001175 OXU24866.1 AJVK01000157 AJVK01000158 AJVK01000159 JH431939 GFDF01009209 JAV04875.1 GL732623 EFX70258.1 JXUM01043842 KQ561377 KXJ78797.1 GFDF01009326 JAV04758.1 GBHO01025127 GBHO01025126 JAG18477.1 JAG18478.1 KQ434889 KZC10220.1 GBRD01014970 GBRD01011697 GBRD01003243 JAG50856.1 NEDP02076705 OWF35828.1 DS235100 EEB11998.1 GFTR01008454 JAW07972.1 AJWK01026643 GG666616 EEN48463.1 CH933807 EDW11294.1 CH940649 EDW63914.1 CH964274 EDW85282.1 CP012523 ALC39746.1 NHOQ01002357 PWA17617.1 CH954177 KQS70574.1 CH480818 EDW52564.1 CM000361 CM002910 EDX04667.1 KMY89702.1 EDV58922.1 HAED01008057 SBQ94269.1 HAEE01004298 SBR24318.1 HAEH01002819 HAEI01009100 SBS06829.1 HAEF01020830 HAEG01001521 SBR61989.1 HADW01011454 HADX01015471 SBP12854.1 HADZ01012048 HAEA01008323 SBQ36803.1 HADY01003789 HAEJ01006774 SBP42274.1 OUUW01000004 SPP79475.1 CH902620 EDV32061.2 HAEB01001958 HAEC01012373 SBQ80590.1 AE014134 BT053681 AAF53072.1 ACK77596.1 CH379060 EAL33855.1 AERX01007738 CH479180 EDW29119.1 AF145632 AAD38607.1 KQ041602 KKF24394.1 AYCK01014652 CM000157 EDW88048.1 JARO02002360 KPP72851.1
ADN88179.1 KQ459601 KPI93983.1 AGBW02013052 OWR44040.1 JTDY01000232 KOB78149.1 GEDC01027435 GEDC01012390 JAS09863.1 JAS24908.1 KQ460855 KPJ11901.1 KQ971343 KYB27482.1 GDKW01000507 JAI56088.1 GALX01003399 JAB65067.1 KK852846 KDR15104.1 KZ288391 PBC26350.1 KQ435770 KOX75180.1 ACPB03011142 KQ763867 OAD54693.1 KQ982769 KYQ50837.1 KQ414786 KOC60801.1 GL445914 EFN88845.1 KK107455 EZA50425.1 LBMM01000533 KMQ98115.1 GL435569 EFN73120.1 KQ978023 KYM97902.1 KQ980636 KYN14959.1 GL888182 EGI65695.1 KQ981490 KYN41044.1 KQ976401 KYM92554.1 ADTU01013223 UFQS01000295 UFQT01000295 SSX02575.1 SSX22949.1 CH477233 EAT46757.1 AJWK01007292 GDHC01007247 JAQ11382.1 GBBM01000427 JAC34991.1 CVRI01000055 CRL01594.1 GANO01001647 JAB58224.1 GFDF01009210 JAV04874.1 NNAY01001175 OXU24866.1 AJVK01000157 AJVK01000158 AJVK01000159 JH431939 GFDF01009209 JAV04875.1 GL732623 EFX70258.1 JXUM01043842 KQ561377 KXJ78797.1 GFDF01009326 JAV04758.1 GBHO01025127 GBHO01025126 JAG18477.1 JAG18478.1 KQ434889 KZC10220.1 GBRD01014970 GBRD01011697 GBRD01003243 JAG50856.1 NEDP02076705 OWF35828.1 DS235100 EEB11998.1 GFTR01008454 JAW07972.1 AJWK01026643 GG666616 EEN48463.1 CH933807 EDW11294.1 CH940649 EDW63914.1 CH964274 EDW85282.1 CP012523 ALC39746.1 NHOQ01002357 PWA17617.1 CH954177 KQS70574.1 CH480818 EDW52564.1 CM000361 CM002910 EDX04667.1 KMY89702.1 EDV58922.1 HAED01008057 SBQ94269.1 HAEE01004298 SBR24318.1 HAEH01002819 HAEI01009100 SBS06829.1 HAEF01020830 HAEG01001521 SBR61989.1 HADW01011454 HADX01015471 SBP12854.1 HADZ01012048 HAEA01008323 SBQ36803.1 HADY01003789 HAEJ01006774 SBP42274.1 OUUW01000004 SPP79475.1 CH902620 EDV32061.2 HAEB01001958 HAEC01012373 SBQ80590.1 AE014134 BT053681 AAF53072.1 ACK77596.1 CH379060 EAL33855.1 AERX01007738 CH479180 EDW29119.1 AF145632 AAD38607.1 KQ041602 KKF24394.1 AYCK01014652 CM000157 EDW88048.1 JARO02002360 KPP72851.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000053240
+ More
UP000007266 UP000005203 UP000027135 UP000242457 UP000053105 UP000015103 UP000075809 UP000053825 UP000008237 UP000053097 UP000036403 UP000000311 UP000078542 UP000078492 UP000007755 UP000078541 UP000078540 UP000005205 UP000008820 UP000075885 UP000092461 UP000183832 UP000215335 UP000092462 UP000000305 UP000069940 UP000249989 UP000076502 UP000242188 UP000009046 UP000001554 UP000009192 UP000261560 UP000008792 UP000265080 UP000261380 UP000007798 UP000002852 UP000092553 UP000192224 UP000261400 UP000008711 UP000001292 UP000000304 UP000018467 UP000265200 UP000265180 UP000261420 UP000261360 UP000265160 UP000268350 UP000007801 UP000001038 UP000000803 UP000261460 UP000001819 UP000261500 UP000005207 UP000192221 UP000008744 UP000261480 UP000028760 UP000002282 UP000034805 UP000242638
UP000007266 UP000005203 UP000027135 UP000242457 UP000053105 UP000015103 UP000075809 UP000053825 UP000008237 UP000053097 UP000036403 UP000000311 UP000078542 UP000078492 UP000007755 UP000078541 UP000078540 UP000005205 UP000008820 UP000075885 UP000092461 UP000183832 UP000215335 UP000092462 UP000000305 UP000069940 UP000249989 UP000076502 UP000242188 UP000009046 UP000001554 UP000009192 UP000261560 UP000008792 UP000265080 UP000261380 UP000007798 UP000002852 UP000092553 UP000192224 UP000261400 UP000008711 UP000001292 UP000000304 UP000018467 UP000265200 UP000265180 UP000261420 UP000261360 UP000265160 UP000268350 UP000007801 UP000001038 UP000000803 UP000261460 UP000001819 UP000261500 UP000005207 UP000192221 UP000008744 UP000261480 UP000028760 UP000002282 UP000034805 UP000242638
Interpro
IPR023299
ATPase_P-typ_cyto_dom_N
+ More
IPR001757 P_typ_ATPase
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR006544 P-type_TPase_V
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR018303 ATPase_P-typ_P_site
IPR001734 Na/solute_symporter
IPR038377 Na/Glc_symporter_sf
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR016187 CTDL_fold
IPR001757 P_typ_ATPase
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR006544 P-type_TPase_V
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR018303 ATPase_P-typ_P_site
IPR001734 Na/solute_symporter
IPR038377 Na/Glc_symporter_sf
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR016187 CTDL_fold
SUPFAM
ProteinModelPortal
H9J9G9
A0A2H1WHX9
A0A2A4JAF7
E2JE29
A0A194PLW6
A0A212ERB9
+ More
A0A0L7LRZ6 A0A1B6DGW9 A0A194R7Q5 A0A139WHP6 A0A0N7Z9H8 V5GTV3 A0A088ALT2 A0A067QZV5 A0A2A3E3S8 A0A0M9A125 T1HIV5 A0A310SHT1 A0A151WSR6 A0A0L7QQK9 E2B656 A0A026W2W9 A0A0J7P0G2 E2A0B7 A0A195CAR3 A0A151J0E3 F4WJB7 A0A195FM34 A0A195BVA2 A0A158NEA4 A0A336KH29 Q17JE6 A0A182PMM5 A0A1B0CD08 A0A146LXI2 A0A023GMC8 A0A1J1IPE0 U5EKZ4 A0A1L8DEW8 A0A232F228 A0A1B0CYG6 T1J7Q5 A0A1L8DEQ9 E9HDF2 A0A182G5Q4 A0A1L8DE88 A0A0A9XCM8 A0A154PG96 A0A0K8SC29 A0A210PH55 E0VF24 A0A224XHB2 A0A1B0CSV1 C3ZFY8 B4KI12 A0A3B3BHL8 B4LTE4 A0A3P8SCX5 A0A3B5LYP7 B4NLP3 M4AG25 A0A0M4E649 A0A315V3K5 A0A1W5ABI2 A0A3B5AQC8 A0A0Q5WJS8 B4HX19 B4QAB2 A0A3B1KED8 B3N4L0 A0A3P9HZV1 A0A3P9JXX7 A0A1A8IBQ6 A0A3B4UT60 A0A1A8JW57 A0A1A8RNK4 A0A1A8MZQ8 A0A1A7X4V3 A0A1A8DT76 A0A3B4WE62 A0A3P9C8N9 A0A1A7ZHX3 A0A3B0JBQ6 B3MNF3 A0A3B3HYE3 A0A1A8HD03 Q9VKJ6 A0A3B4FLV9 Q29M40 A0A3B3W2A6 I3J0F7 A0A1W4W523 B4G9T9 A0A3B3X723 Q9Y139 A0A0F8B578 A0A087XMC1 B4P173 A0A0P7UU04 A0A3P9NDK9
A0A0L7LRZ6 A0A1B6DGW9 A0A194R7Q5 A0A139WHP6 A0A0N7Z9H8 V5GTV3 A0A088ALT2 A0A067QZV5 A0A2A3E3S8 A0A0M9A125 T1HIV5 A0A310SHT1 A0A151WSR6 A0A0L7QQK9 E2B656 A0A026W2W9 A0A0J7P0G2 E2A0B7 A0A195CAR3 A0A151J0E3 F4WJB7 A0A195FM34 A0A195BVA2 A0A158NEA4 A0A336KH29 Q17JE6 A0A182PMM5 A0A1B0CD08 A0A146LXI2 A0A023GMC8 A0A1J1IPE0 U5EKZ4 A0A1L8DEW8 A0A232F228 A0A1B0CYG6 T1J7Q5 A0A1L8DEQ9 E9HDF2 A0A182G5Q4 A0A1L8DE88 A0A0A9XCM8 A0A154PG96 A0A0K8SC29 A0A210PH55 E0VF24 A0A224XHB2 A0A1B0CSV1 C3ZFY8 B4KI12 A0A3B3BHL8 B4LTE4 A0A3P8SCX5 A0A3B5LYP7 B4NLP3 M4AG25 A0A0M4E649 A0A315V3K5 A0A1W5ABI2 A0A3B5AQC8 A0A0Q5WJS8 B4HX19 B4QAB2 A0A3B1KED8 B3N4L0 A0A3P9HZV1 A0A3P9JXX7 A0A1A8IBQ6 A0A3B4UT60 A0A1A8JW57 A0A1A8RNK4 A0A1A8MZQ8 A0A1A7X4V3 A0A1A8DT76 A0A3B4WE62 A0A3P9C8N9 A0A1A7ZHX3 A0A3B0JBQ6 B3MNF3 A0A3B3HYE3 A0A1A8HD03 Q9VKJ6 A0A3B4FLV9 Q29M40 A0A3B3W2A6 I3J0F7 A0A1W4W523 B4G9T9 A0A3B3X723 Q9Y139 A0A0F8B578 A0A087XMC1 B4P173 A0A0P7UU04 A0A3P9NDK9
PDB
3WGV
E-value=4.16128e-20,
Score=246
Ontologies
GO
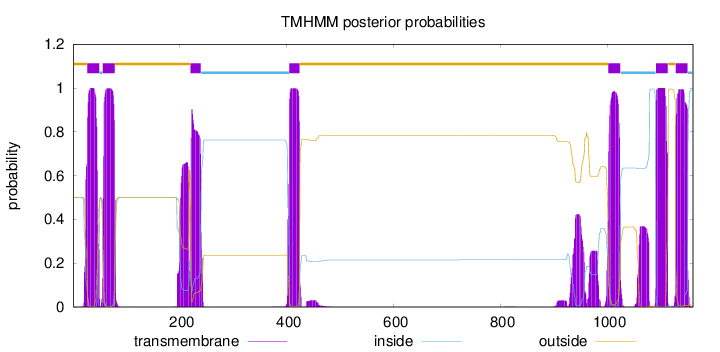
Topology
Subcellular location
Membrane
Length:
1160
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
185.29739
Exp number, first 60 AAs:
26.78265
Total prob of N-in:
0.49873
POSSIBLE N-term signal
sequence
outside
1 - 26
TMhelix
27 - 49
inside
50 - 55
TMhelix
56 - 78
outside
79 - 219
TMhelix
220 - 239
inside
240 - 404
TMhelix
405 - 424
outside
425 - 1001
TMhelix
1002 - 1024
inside
1025 - 1090
TMhelix
1091 - 1113
outside
1114 - 1127
TMhelix
1128 - 1150
inside
1151 - 1160
Population Genetic Test Statistics
Pi
182.064312
Theta
165.994095
Tajima's D
0.183068
CLR
0.283247
CSRT
0.427978601069947
Interpretation
Uncertain
