Gene
KWMTBOMO01804
Pre Gene Modal
BGIBMGA006161
Annotation
PREDICTED:_Golgi_apparatus_protein_1_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 2.495
Sequence
CDS
ATGTCAACTAAAGTCTTCACGACAGTTTGGGTAATAACGGCATGTTTAGGAGTTTTGAAAAATGTGCATTGCCGAAAATCGGATGATAACTCCTACAAAAACTATTTATTCAGCTGGGACGCTGGTGAATGCGCGCTGTTTCATAAATTATGTTCTCACTTTGACGATCATTTCGCTTTGCTGACATGTTCATTACATTCGTCCTATGATAAGACCCAAATACCACCAAGTTGTCAGCACAAGTTATGGGTTCAGCTGTCATACTTAATCAACAACTCATTCTTGTTTCATACACTAAAAGACCCATGTCAAGAAGATACAGAAATATCGAGTTGTTTTGATCCTAATGTCTTAACTGTAGATTGTATATTAAAAAAAAAGCCTAGTGCGACAGTGAAAGTTTGCTGGAGAATGATAAATAAAATAGAATCTCTATTATTTAATGACTGGCAAATTACTACAAACTTCATTTCTAAATGTTTTGATGATGTAAATACTCATACTTGTGGTAAAATTCCACCTGATCCTAAGAGTCTTTCGCAAACAGAGACATTGAAATGTCTTCAAAACTATGAACAATATGTCACTGCAGATTGTAAAGCAGAGATCACTGTACTATATGAAATGAGATACAGTAATTTACAGTTAGATAAAATTGTATTTGCTGCCTGTAATGTAGAACAAAATAGTTTCTGTCGTGATGAAATACCAGGATCATGGTTAATGTATAAATGTCTCCTGAGACATAAGTATGAAAATGGGGTTCGTAAAAAATGTCAAGATCAGCTCTTCTATGATCAAAAAAATATAGCCATGAATTATAAAATGAGTCGAGGTTTAGTAAAGGCATGCAGAGAAGACATTAGAAAGTTTCATTGTCGAAAAGGCATTGTTGAAGACAAAGATGTGAGATTAGCACAAATTTTACTCTGCTTAGAGAGTAACATACAAAATGATACGAAAAGCATTTCGTCTGAGTGTACAGTGGAAATGATTGATCATAGAAAAATGTTGATGGAAGATTATAGACTGTCACCAGAGTTGATGCAGAATTGTGCCAATGACATTACTATGCTATGTAGAGGAATTGAAGCTGGTGGTAAAACTATTCACTGCTTAATGGACCATGCAAGACCTAGGAGAAGAAAAGACAGAAGAATAAGTTTAGAATGTCAGAAGTCATTGGAAATGTTAGTACAGGAAACAAATCCAGGTGAAGATTGGAGAGTTGATCCTGTTTTAAGGGCAGCCTGTAAACCTGTTGTAGATAAAGCATGTGGAGAAATAGCTGGTGGTAATGGCAGAGTCATGTCATGTCTAATGGAAAAAATTCATACAAAAGCAATGACTGATGAATGTGAGGTGGCATTATTGCAAATACAGTACTTCATATCAAGAGATTTTAAGCTTGATCCTCAGTTATATAAAGCTTGTAAGTATGATGCTGTTACTACTTGCAAGGCTAAGTTACAATGGGCTGATGCTAGTGAACATCATTCTGAAAAAGATCCTTTAATATTTCCATGCTTATATAATTATGCTTACAACTCTGAATTGAGAGGCAGATTGCGACTAGCCTGTGAGGAACAAGTGAAGAGGGTAATGAGGCAGAGAGCATCAAGTGTTGATTTAATGCCACAAATTGAGGACTTTTGCATGGATGACTTAGCCAATTTTTGTCTTGAGAATACTGGAAAAGGGGAAGAAATTTTATGTTTACAAAGTAAAATTAAAGAGCTTTCTCCAAAGTGTAAAGATATTGTGTCTAATTTCACTGAAACTCAAAGTGGACACATAGAACTAAATGCAGTTATCAATACAAACTGTCGAGGTATTACTGAAAAACTATGTGCATCTGAACTACAAAGTAAAAAAGAAGAAGATGATGTTCTCGAGTGCCTAATACTTCATAAAAATCATCCAGATGTAAAAGCTAATGTGAAATGCAGAGCTGCCATTGAACACGAACAATTGATCTCATTAAAGAATTATAGATTCACAACCAAGTTTAAGAATGCTTGTAAATATAATGTTGACAGATTTTGTCCCAAAGCACAAACAAAAAACCAAGTTGTATCATGTTTGAGCGAGATAGTAAGAAATGATACAATTAACAGACGTAAACATACAATAGTCAAGGAGTGTCGTCAACAGCTAAGAAGTCAATTATTCCAACAAAAGGAAAATATAGATTTAGACCCGGATCTGAAGGAAGCTTGTAAAAAAGATCTTGAAATATTATGCCCTTCTATTACACATGGTGAGGCTGCAGCCTTAGAGTGCCTTCAGACTTCCAAAGGAAAATTGAGTCAAAAATGTAGGAAGGTATTGTTTGTAGTGAAGAAACAAGAATTTACAGACAGCGGAATTGATTACCATTTGATAAAAACATGCAATGACATGATAGATAAATTCTGCCACAACACAGAACCCTCAAAGATTTTTGACTGTTTGAAAACACATCATCAGAGCCTTGATTTCGAGAAAAATTGTAAAGTTGTTGTGGTGAACCGAATGATAGAACAGAATACAGATTACAGATTTAACATAAATTTGGTCAATTCATGTAAATCTGATATTAATGCACATTGCAGTGAAATATTAGCTAATGAACCTCAAGATGTAGAACTACAAGGAAAAGTATTGTATTGCCTAAAAGAAAAGTTTAGGGAATCAAAATTAACGACAGATTGTGAAAATGAAGTGGCTAATATTTTGAAGGAGCAGGCATTGAATTATCGTCTTGATCCATTGCTTGAGAAGCTGTGTCAAGCTGAAATTCAAACGATATGTTCAGTGCAAAACGATATCACTATTTCGGATGGACAGGTGGAGGAATGCCTCAAGAATGCCTTATTAAAAGACAAAATCGTATCAGCCGAATGCTCCCGTGAAATAGCTAAGTTGATAGAACAAACCGAAGTCGACATCGAGGCTGATCCGTTGTTAGAAAGAGCCTGTGCTTTCGATCTGTTGAAGTATTGTAAAGATCTAGAACATGGTGCTGGGAGACGTCTGAAATGTTTAAAAATTATATCAAATGATGGAACACGGAAGTTGGAGGCAGCATGTGAAAAGGAACTCACAAGTAGATTAGAAATGTATAGATTTGTAGCAGCTAAATTTGAAACACTGGGAGATGCATAA
Protein
MSTKVFTTVWVITACLGVLKNVHCRKSDDNSYKNYLFSWDAGECALFHKLCSHFDDHFALLTCSLHSSYDKTQIPPSCQHKLWVQLSYLINNSFLFHTLKDPCQEDTEISSCFDPNVLTVDCILKKKPSATVKVCWRMINKIESLLFNDWQITTNFISKCFDDVNTHTCGKIPPDPKSLSQTETLKCLQNYEQYVTADCKAEITVLYEMRYSNLQLDKIVFAACNVEQNSFCRDEIPGSWLMYKCLLRHKYENGVRKKCQDQLFYDQKNIAMNYKMSRGLVKACREDIRKFHCRKGIVEDKDVRLAQILLCLESNIQNDTKSISSECTVEMIDHRKMLMEDYRLSPELMQNCANDITMLCRGIEAGGKTIHCLMDHARPRRRKDRRISLECQKSLEMLVQETNPGEDWRVDPVLRAACKPVVDKACGEIAGGNGRVMSCLMEKIHTKAMTDECEVALLQIQYFISRDFKLDPQLYKACKYDAVTTCKAKLQWADASEHHSEKDPLIFPCLYNYAYNSELRGRLRLACEEQVKRVMRQRASSVDLMPQIEDFCMDDLANFCLENTGKGEEILCLQSKIKELSPKCKDIVSNFTETQSGHIELNAVINTNCRGITEKLCASELQSKKEEDDVLECLILHKNHPDVKANVKCRAAIEHEQLISLKNYRFTTKFKNACKYNVDRFCPKAQTKNQVVSCLSEIVRNDTINRRKHTIVKECRQQLRSQLFQQKENIDLDPDLKEACKKDLEILCPSITHGEAAALECLQTSKGKLSQKCRKVLFVVKKQEFTDSGIDYHLIKTCNDMIDKFCHNTEPSKIFDCLKTHHQSLDFEKNCKVVVVNRMIEQNTDYRFNINLVNSCKSDINAHCSEILANEPQDVELQGKVLYCLKEKFRESKLTTDCENEVANILKEQALNYRLDPLLEKLCQAEIQTICSVQNDITISDGQVEECLKNALLKDKIVSAECSREIAKLIEQTEVDIEADPLLERACAFDLLKYCKDLEHGAGRRLKCLKIISNDGTRKLEAACEKELTSRLEMYRFVAAKFETLGDA
Summary
Uniprot
H9J9G8
A0A2A4J8D8
A0A2H1WHZ0
A0A2A4J8I0
A0A2A4J9F1
A0A194R302
+ More
A0A194PS82 A0A212F7X7 A0A0L7LRR6 U5EST3 A0A1Q3FDG2 B0X7U9 A0A1Q3FG99 Q174F8 A0A1S4FF42 A0A1Q3FGN0 A0A1Q3FEF1 A0A1Q3FDQ9 A0A182H9E8 A0A2J7RB29 A0A084VS89 A0A067R9F6 A0A336LUL6 A0A182YBC8 A0A182NQJ3 A0A182P9L1 A0A182HKK4 A0A182ILE1 A0A3F2YUZ0 Q7PXB5 A0A182QDE3 A0A182X9M0 A0A182TE04 A0A182UWL7 A0A182JTB0 A0A182VVS2 A0A182MDQ0 W5J316 A0A2M4BB87 A0A2M4BB69 A0A2M4BB62 A0A2M4A886 A0A182RNK8 A0A1L8DVX6 A0A1L8DVH7 A0A1L8DVI9 A0A1L8DVJ1 A0A1L8DVK3 A0A1L8DVJ8 A0A2M4BBR4 A0A2M4BB75 A0A2M4BBA2 A0A1B6EC31 A0A182FSQ4 A0A1L8DVF2 A0A2M3ZDX2 E2AIP6 A0A2M3ZGU5 A0A026WUB3 A0A151XHS3 A0A154PF01 A0A2M4A9G9 A0A151IL20 E9J083 A0A195FHK8 A0A1B0CLR1 A0A158ND65 F4X7A1 E2C6H1 A0A310SRV0 A0A195DJ67 A0A2M3Z3S2 A0A2P8ZID3 A0A1W6EVR7 A0A195B0W4 A0A088AD84 K7IXM7 A0A0J7L8F0 A0A1W4WSH8 A0A232F0E4 A0A0C9QL54 A0A0C9PJY9 A0A1Y1M578 A0A1B6FJY6 A0A1W4X394 A0A1B6HQR9 A0A1B6KZZ0 A0A0A1XH87 A0A1W4UEK4 A0A0M4EJH7 W8BDX5 A0A1J1HZV2 A0A034VNQ1 A0A0A1XPK2 A0A1W4URL8 B4IWW2 A0A0K8UKZ2 W8C184 A0A0M8ZXG4 A0A034VR47
A0A194PS82 A0A212F7X7 A0A0L7LRR6 U5EST3 A0A1Q3FDG2 B0X7U9 A0A1Q3FG99 Q174F8 A0A1S4FF42 A0A1Q3FGN0 A0A1Q3FEF1 A0A1Q3FDQ9 A0A182H9E8 A0A2J7RB29 A0A084VS89 A0A067R9F6 A0A336LUL6 A0A182YBC8 A0A182NQJ3 A0A182P9L1 A0A182HKK4 A0A182ILE1 A0A3F2YUZ0 Q7PXB5 A0A182QDE3 A0A182X9M0 A0A182TE04 A0A182UWL7 A0A182JTB0 A0A182VVS2 A0A182MDQ0 W5J316 A0A2M4BB87 A0A2M4BB69 A0A2M4BB62 A0A2M4A886 A0A182RNK8 A0A1L8DVX6 A0A1L8DVH7 A0A1L8DVI9 A0A1L8DVJ1 A0A1L8DVK3 A0A1L8DVJ8 A0A2M4BBR4 A0A2M4BB75 A0A2M4BBA2 A0A1B6EC31 A0A182FSQ4 A0A1L8DVF2 A0A2M3ZDX2 E2AIP6 A0A2M3ZGU5 A0A026WUB3 A0A151XHS3 A0A154PF01 A0A2M4A9G9 A0A151IL20 E9J083 A0A195FHK8 A0A1B0CLR1 A0A158ND65 F4X7A1 E2C6H1 A0A310SRV0 A0A195DJ67 A0A2M3Z3S2 A0A2P8ZID3 A0A1W6EVR7 A0A195B0W4 A0A088AD84 K7IXM7 A0A0J7L8F0 A0A1W4WSH8 A0A232F0E4 A0A0C9QL54 A0A0C9PJY9 A0A1Y1M578 A0A1B6FJY6 A0A1W4X394 A0A1B6HQR9 A0A1B6KZZ0 A0A0A1XH87 A0A1W4UEK4 A0A0M4EJH7 W8BDX5 A0A1J1HZV2 A0A034VNQ1 A0A0A1XPK2 A0A1W4URL8 B4IWW2 A0A0K8UKZ2 W8C184 A0A0M8ZXG4 A0A034VR47
Pubmed
EMBL
BABH01004811
NWSH01002391
PCG68387.1
ODYU01008780
SOQ52678.1
PCG68385.1
+ More
PCG68386.1 KQ460855 KPJ11899.1 KQ459601 KPI93985.1 AGBW02009822 OWR49845.1 JTDY01000232 KOB78150.1 GANO01002219 JAB57652.1 GFDL01009440 JAV25605.1 DS232464 EDS42168.1 GFDL01008472 JAV26573.1 CH477412 EAT41434.1 GFDL01008338 JAV26707.1 GFDL01009112 JAV25933.1 GFDL01009348 JAV25697.1 JXUM01121069 JXUM01121070 JXUM01121071 JXUM01121072 JXUM01121073 KQ566459 KXJ70119.1 NEVH01006564 PNF38032.1 ATLV01015877 KE525039 KFB40833.1 KK852804 KDR16244.1 UFQT01000117 SSX20369.1 APCN01000836 AAAB01008987 EAA01189.4 AXCN02001519 AXCM01000656 ADMH02002125 ETN58727.1 GGFJ01001131 MBW50272.1 GGFJ01001136 MBW50277.1 GGFJ01001135 MBW50276.1 GGFK01003693 MBW37014.1 GFDF01003639 JAV10445.1 GFDF01003640 JAV10444.1 GFDF01003636 JAV10448.1 GFDF01003637 JAV10447.1 GFDF01003641 JAV10443.1 GFDF01003638 JAV10446.1 GGFJ01001117 MBW50258.1 GGFJ01001121 MBW50262.1 GGFJ01001120 MBW50261.1 GEDC01001851 JAS35447.1 GFDF01003666 JAV10418.1 GGFM01006003 MBW26754.1 GL439852 EFN66701.1 GGFM01006978 MBW27729.1 KK107102 QOIP01000005 EZA59620.1 RLU22520.1 KQ982116 KYQ59952.1 KQ434889 KZC10387.1 GGFK01004098 MBW37419.1 KQ977146 KYN05315.1 GL767333 EFZ13759.1 KQ981560 KYN39873.1 AJWK01017543 ADTU01012434 GL888828 EGI57713.1 GL453156 EFN76453.1 KQ760551 OAD59812.1 KQ980800 KYN12933.1 GGFM01002416 MBW23167.1 PYGN01000048 PSN56256.1 KY563414 ARK19823.1 KQ976690 KYM78101.1 AAZX01000042 LBMM01000218 KMR04347.1 NNAY01001405 OXU24082.1 GBYB01001277 JAG71044.1 GBYB01001278 JAG71045.1 GEZM01040038 JAV80979.1 GECZ01019251 GECZ01018664 JAS50518.1 JAS51105.1 GECU01030674 GECU01000778 JAS77032.1 JAT06929.1 GEBQ01022948 JAT17029.1 GBXI01003990 JAD10302.1 CP012525 ALC45102.1 GAMC01007100 JAB99455.1 CVRI01000023 CRK92078.1 GAKP01014001 JAC44951.1 GBXI01001336 JAD12956.1 CH916366 EDV97363.1 GDHF01025121 JAI27193.1 GAMC01007099 JAB99456.1 KQ435811 KOX72907.1 GAKP01014003 JAC44949.1
PCG68386.1 KQ460855 KPJ11899.1 KQ459601 KPI93985.1 AGBW02009822 OWR49845.1 JTDY01000232 KOB78150.1 GANO01002219 JAB57652.1 GFDL01009440 JAV25605.1 DS232464 EDS42168.1 GFDL01008472 JAV26573.1 CH477412 EAT41434.1 GFDL01008338 JAV26707.1 GFDL01009112 JAV25933.1 GFDL01009348 JAV25697.1 JXUM01121069 JXUM01121070 JXUM01121071 JXUM01121072 JXUM01121073 KQ566459 KXJ70119.1 NEVH01006564 PNF38032.1 ATLV01015877 KE525039 KFB40833.1 KK852804 KDR16244.1 UFQT01000117 SSX20369.1 APCN01000836 AAAB01008987 EAA01189.4 AXCN02001519 AXCM01000656 ADMH02002125 ETN58727.1 GGFJ01001131 MBW50272.1 GGFJ01001136 MBW50277.1 GGFJ01001135 MBW50276.1 GGFK01003693 MBW37014.1 GFDF01003639 JAV10445.1 GFDF01003640 JAV10444.1 GFDF01003636 JAV10448.1 GFDF01003637 JAV10447.1 GFDF01003641 JAV10443.1 GFDF01003638 JAV10446.1 GGFJ01001117 MBW50258.1 GGFJ01001121 MBW50262.1 GGFJ01001120 MBW50261.1 GEDC01001851 JAS35447.1 GFDF01003666 JAV10418.1 GGFM01006003 MBW26754.1 GL439852 EFN66701.1 GGFM01006978 MBW27729.1 KK107102 QOIP01000005 EZA59620.1 RLU22520.1 KQ982116 KYQ59952.1 KQ434889 KZC10387.1 GGFK01004098 MBW37419.1 KQ977146 KYN05315.1 GL767333 EFZ13759.1 KQ981560 KYN39873.1 AJWK01017543 ADTU01012434 GL888828 EGI57713.1 GL453156 EFN76453.1 KQ760551 OAD59812.1 KQ980800 KYN12933.1 GGFM01002416 MBW23167.1 PYGN01000048 PSN56256.1 KY563414 ARK19823.1 KQ976690 KYM78101.1 AAZX01000042 LBMM01000218 KMR04347.1 NNAY01001405 OXU24082.1 GBYB01001277 JAG71044.1 GBYB01001278 JAG71045.1 GEZM01040038 JAV80979.1 GECZ01019251 GECZ01018664 JAS50518.1 JAS51105.1 GECU01030674 GECU01000778 JAS77032.1 JAT06929.1 GEBQ01022948 JAT17029.1 GBXI01003990 JAD10302.1 CP012525 ALC45102.1 GAMC01007100 JAB99455.1 CVRI01000023 CRK92078.1 GAKP01014001 JAC44951.1 GBXI01001336 JAD12956.1 CH916366 EDV97363.1 GDHF01025121 JAI27193.1 GAMC01007099 JAB99456.1 KQ435811 KOX72907.1 GAKP01014003 JAC44949.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000002320 UP000008820 UP000069940 UP000249989 UP000235965 UP000030765 UP000027135 UP000076408 UP000075884 UP000075885 UP000075840 UP000075880 UP000075882 UP000007062 UP000075886 UP000076407 UP000075902 UP000075903 UP000075881 UP000075920 UP000075883 UP000000673 UP000075900 UP000069272 UP000000311 UP000053097 UP000279307 UP000075809 UP000076502 UP000078542 UP000078541 UP000092461 UP000005205 UP000007755 UP000008237 UP000078492 UP000245037 UP000078540 UP000005203 UP000002358 UP000036403 UP000192223 UP000215335 UP000192221 UP000092553 UP000183832 UP000001070 UP000053105
UP000002320 UP000008820 UP000069940 UP000249989 UP000235965 UP000030765 UP000027135 UP000076408 UP000075884 UP000075885 UP000075840 UP000075880 UP000075882 UP000007062 UP000075886 UP000076407 UP000075902 UP000075903 UP000075881 UP000075920 UP000075883 UP000000673 UP000075900 UP000069272 UP000000311 UP000053097 UP000279307 UP000075809 UP000076502 UP000078542 UP000078541 UP000092461 UP000005205 UP000007755 UP000008237 UP000078492 UP000245037 UP000078540 UP000005203 UP000002358 UP000036403 UP000192223 UP000215335 UP000192221 UP000092553 UP000183832 UP000001070 UP000053105
PRIDE
Interpro
SUPFAM
SSF56784
SSF56784
Gene 3D
ProteinModelPortal
H9J9G8
A0A2A4J8D8
A0A2H1WHZ0
A0A2A4J8I0
A0A2A4J9F1
A0A194R302
+ More
A0A194PS82 A0A212F7X7 A0A0L7LRR6 U5EST3 A0A1Q3FDG2 B0X7U9 A0A1Q3FG99 Q174F8 A0A1S4FF42 A0A1Q3FGN0 A0A1Q3FEF1 A0A1Q3FDQ9 A0A182H9E8 A0A2J7RB29 A0A084VS89 A0A067R9F6 A0A336LUL6 A0A182YBC8 A0A182NQJ3 A0A182P9L1 A0A182HKK4 A0A182ILE1 A0A3F2YUZ0 Q7PXB5 A0A182QDE3 A0A182X9M0 A0A182TE04 A0A182UWL7 A0A182JTB0 A0A182VVS2 A0A182MDQ0 W5J316 A0A2M4BB87 A0A2M4BB69 A0A2M4BB62 A0A2M4A886 A0A182RNK8 A0A1L8DVX6 A0A1L8DVH7 A0A1L8DVI9 A0A1L8DVJ1 A0A1L8DVK3 A0A1L8DVJ8 A0A2M4BBR4 A0A2M4BB75 A0A2M4BBA2 A0A1B6EC31 A0A182FSQ4 A0A1L8DVF2 A0A2M3ZDX2 E2AIP6 A0A2M3ZGU5 A0A026WUB3 A0A151XHS3 A0A154PF01 A0A2M4A9G9 A0A151IL20 E9J083 A0A195FHK8 A0A1B0CLR1 A0A158ND65 F4X7A1 E2C6H1 A0A310SRV0 A0A195DJ67 A0A2M3Z3S2 A0A2P8ZID3 A0A1W6EVR7 A0A195B0W4 A0A088AD84 K7IXM7 A0A0J7L8F0 A0A1W4WSH8 A0A232F0E4 A0A0C9QL54 A0A0C9PJY9 A0A1Y1M578 A0A1B6FJY6 A0A1W4X394 A0A1B6HQR9 A0A1B6KZZ0 A0A0A1XH87 A0A1W4UEK4 A0A0M4EJH7 W8BDX5 A0A1J1HZV2 A0A034VNQ1 A0A0A1XPK2 A0A1W4URL8 B4IWW2 A0A0K8UKZ2 W8C184 A0A0M8ZXG4 A0A034VR47
A0A194PS82 A0A212F7X7 A0A0L7LRR6 U5EST3 A0A1Q3FDG2 B0X7U9 A0A1Q3FG99 Q174F8 A0A1S4FF42 A0A1Q3FGN0 A0A1Q3FEF1 A0A1Q3FDQ9 A0A182H9E8 A0A2J7RB29 A0A084VS89 A0A067R9F6 A0A336LUL6 A0A182YBC8 A0A182NQJ3 A0A182P9L1 A0A182HKK4 A0A182ILE1 A0A3F2YUZ0 Q7PXB5 A0A182QDE3 A0A182X9M0 A0A182TE04 A0A182UWL7 A0A182JTB0 A0A182VVS2 A0A182MDQ0 W5J316 A0A2M4BB87 A0A2M4BB69 A0A2M4BB62 A0A2M4A886 A0A182RNK8 A0A1L8DVX6 A0A1L8DVH7 A0A1L8DVI9 A0A1L8DVJ1 A0A1L8DVK3 A0A1L8DVJ8 A0A2M4BBR4 A0A2M4BB75 A0A2M4BBA2 A0A1B6EC31 A0A182FSQ4 A0A1L8DVF2 A0A2M3ZDX2 E2AIP6 A0A2M3ZGU5 A0A026WUB3 A0A151XHS3 A0A154PF01 A0A2M4A9G9 A0A151IL20 E9J083 A0A195FHK8 A0A1B0CLR1 A0A158ND65 F4X7A1 E2C6H1 A0A310SRV0 A0A195DJ67 A0A2M3Z3S2 A0A2P8ZID3 A0A1W6EVR7 A0A195B0W4 A0A088AD84 K7IXM7 A0A0J7L8F0 A0A1W4WSH8 A0A232F0E4 A0A0C9QL54 A0A0C9PJY9 A0A1Y1M578 A0A1B6FJY6 A0A1W4X394 A0A1B6HQR9 A0A1B6KZZ0 A0A0A1XH87 A0A1W4UEK4 A0A0M4EJH7 W8BDX5 A0A1J1HZV2 A0A034VNQ1 A0A0A1XPK2 A0A1W4URL8 B4IWW2 A0A0K8UKZ2 W8C184 A0A0M8ZXG4 A0A034VR47
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.948576
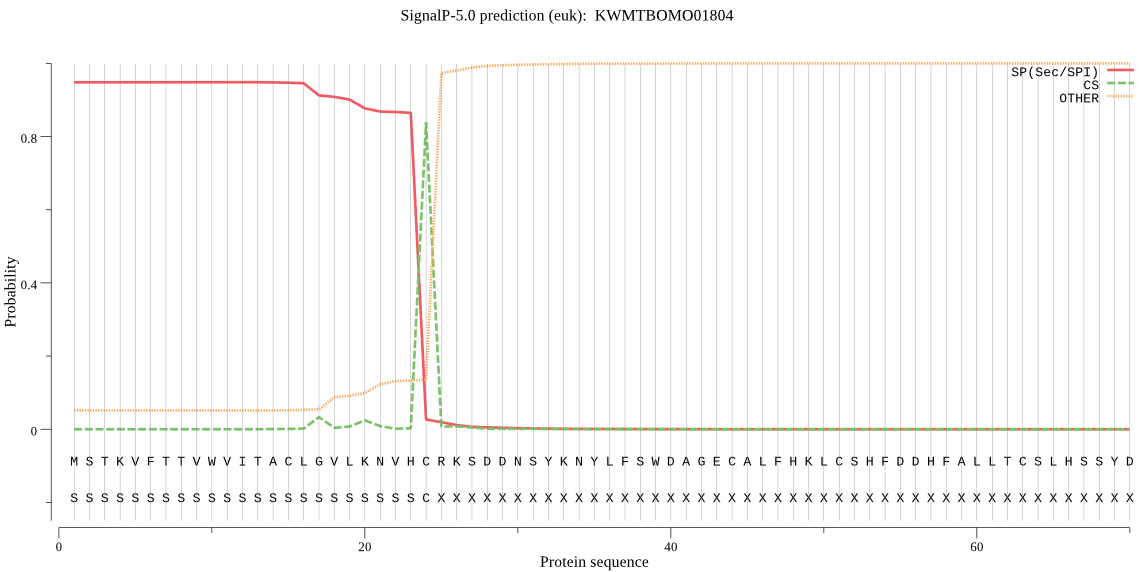
Length:
1048
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14017
Exp number, first 60 AAs:
0.13725
Total prob of N-in:
0.00716
outside
1 - 1048
Population Genetic Test Statistics
Pi
217.857442
Theta
182.771093
Tajima's D
0.502189
CLR
0.282958
CSRT
0.516574171291435
Interpretation
Uncertain
