Gene
KWMTBOMO01803
Pre Gene Modal
BGIBMGA005992
Annotation
PREDICTED:_transmembrane_protein_18_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.641
Sequence
CDS
ATGGAAGAATTCGTTGAAGTTAACCAGATATCTGATTTTGTATCGTACCTAAGAAGTATCGAATGGCGTGACCCCTGGTTAATTGCATTGATATCATTTCACATAATAATTACTGTCACTTGTTTCTCCACCAGGAATTATGGCAATTTTCAAGTTATACTTTTTATCATACTCCTTTTGCTAGTGTATTTTTCGGAGAATATAAATGAGCTTGCTGCAAGAAATTGGGCATTATTTTCAAGACAACAATACTTTGATAGTAAAGGCCTATTTATATCAGTGGTGTTTTCGATTCCTATATTGTTAAACTGCATGATAATGGTGGGATCCTGGTTGTACCAATCTACTCAAATAATGACTAATTTAAAAAAAGCACAGTTAAGGCAGCGCCTCAAGGAGATGAACTCGATAAGAGAACAACAGCAACACGTTAAATCTGATTAA
Protein
MEEFVEVNQISDFVSYLRSIEWRDPWLIALISFHIIITVTCFSTRNYGNFQVILFIILLLLVYFSENINELAARNWALFSRQQYFDSKGLFISVVFSIPILLNCMIMVGSWLYQSTQIMTNLKKAQLRQRLKEMNSIREQQQHVKSD
Summary
Uniprot
A0A1E1VY87
A0A212F7Y0
A0A194R284
A0A194PKM3
H9J900
V5GD49
+ More
A0A023EGS6 Q1HQG4 A0A182N804 A0A182QW26 A0A1Y1L558 A0A2M4C2J1 A0A182UYD0 A0A182TJR8 A0A182LBD2 Q7PQ38 A0A182HP58 A0A182PHE5 T1E923 A0A182LVI4 A0A182WAB7 A0A182XJA4 A0A182FH93 A0A182K137 A0A182YNR7 D6WLC5 A0A1Q3F6A8 B0W5F7 A0A2M3ZCP7 A0A1L8D9U5 A0A182SK74 A0A084W9J7 A0A182ISP2 A0A2M4AU81 W5J9P1 E2AF13 K7J575 A0A232ERW0 A0A088A1C6 A0A2A3ENS3 V9IK05 E2C7C5 A0A182RTQ1 A0A026WEA8 A0A0T6AVM8 B4KR84 A0A1W4XBD6 B4J8V1 U5EGA6 A0A158N9R5 A0A154PQM8 A0A0N0BFD2 T1H6D5 A0A1B6LJ33 A0A3L8DRF0 B4HPJ0 B4QCW6 A0A0A1WZ89 A0A1A9ZB61 A0A1B0C5U5 A0A1B0GBQ7 B3NS06 A1Z903 B4MRI3 A0A0L7QVN5 A0A1B6EVC5 Q4V5Q0 A0A1B6K5P1 A0A195FU22 A0A1W4UHG3 B4MFH0 C1C540 B4P599 B3MEM5 A0A067R500 A0A0M3QUY0 A0A0L7LS04 Q4V5G3 A0A0K8TN79 E0VR95 A0A336KPA6 R4WCV6 A0A1A9YS55 A0A1A9VPQ0 A0A0A9XFQ8 A0A0K8V488 A0A087T0T2 A0A0P4VQ54 A0A2R7WEM3 A0A034WTQ7 A0A1I8PI29 A0A1I8MLV0 A0A1S4EBV3 A0A3Q0ISJ7 T1JB36 A0A224XZU1 A0A069DP02 A0A1B0DM28 B4GGI1 A0A2P6KAL2
A0A023EGS6 Q1HQG4 A0A182N804 A0A182QW26 A0A1Y1L558 A0A2M4C2J1 A0A182UYD0 A0A182TJR8 A0A182LBD2 Q7PQ38 A0A182HP58 A0A182PHE5 T1E923 A0A182LVI4 A0A182WAB7 A0A182XJA4 A0A182FH93 A0A182K137 A0A182YNR7 D6WLC5 A0A1Q3F6A8 B0W5F7 A0A2M3ZCP7 A0A1L8D9U5 A0A182SK74 A0A084W9J7 A0A182ISP2 A0A2M4AU81 W5J9P1 E2AF13 K7J575 A0A232ERW0 A0A088A1C6 A0A2A3ENS3 V9IK05 E2C7C5 A0A182RTQ1 A0A026WEA8 A0A0T6AVM8 B4KR84 A0A1W4XBD6 B4J8V1 U5EGA6 A0A158N9R5 A0A154PQM8 A0A0N0BFD2 T1H6D5 A0A1B6LJ33 A0A3L8DRF0 B4HPJ0 B4QCW6 A0A0A1WZ89 A0A1A9ZB61 A0A1B0C5U5 A0A1B0GBQ7 B3NS06 A1Z903 B4MRI3 A0A0L7QVN5 A0A1B6EVC5 Q4V5Q0 A0A1B6K5P1 A0A195FU22 A0A1W4UHG3 B4MFH0 C1C540 B4P599 B3MEM5 A0A067R500 A0A0M3QUY0 A0A0L7LS04 Q4V5G3 A0A0K8TN79 E0VR95 A0A336KPA6 R4WCV6 A0A1A9YS55 A0A1A9VPQ0 A0A0A9XFQ8 A0A0K8V488 A0A087T0T2 A0A0P4VQ54 A0A2R7WEM3 A0A034WTQ7 A0A1I8PI29 A0A1I8MLV0 A0A1S4EBV3 A0A3Q0ISJ7 T1JB36 A0A224XZU1 A0A069DP02 A0A1B0DM28 B4GGI1 A0A2P6KAL2
Pubmed
22118469
26354079
19121390
24945155
17204158
17510324
+ More
28004739 20966253 12364791 14747013 17210077 25244985 18362917 19820115 24438588 20920257 23761445 20798317 20075255 28648823 24508170 17994087 21347285 30249741 22936249 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24845553 26227816 26369729 20566863 23691247 25401762 26823975 27129103 25348373 25315136 26334808
28004739 20966253 12364791 14747013 17210077 25244985 18362917 19820115 24438588 20920257 23761445 20798317 20075255 28648823 24508170 17994087 21347285 30249741 22936249 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24845553 26227816 26369729 20566863 23691247 25401762 26823975 27129103 25348373 25315136 26334808
EMBL
GDQN01011374
GDQN01006928
JAT79680.1
JAT84126.1
AGBW02009822
OWR49846.1
+ More
KQ460855 KPJ11898.1 KQ459601 KPI93986.1 BABH01004810 GALX01000386 JAB68080.1 GAPW01005589 GEHC01001452 JAC08009.1 JAV46193.1 DQ440480 CH477847 ABF18513.1 EAT35676.1 AXCN02000910 GEZM01070907 JAV66227.1 GGFJ01010391 MBW59532.1 AAAB01008900 EAA09360.5 APCN01002921 GAMD01002238 JAA99352.1 AXCM01001928 KQ971343 EFA03470.1 GFDL01011945 JAV23100.1 DS231842 EDS35277.1 GGFM01005518 MBW26269.1 GFDF01010872 JAV03212.1 ATLV01021826 KE525324 KFB46891.1 GGFK01011016 MBW44337.1 ADMH02002078 ETN59464.1 GL438984 EFN67968.1 AAZX01000344 NNAY01002545 OXU21090.1 KZ288215 PBC32779.1 JR051309 AEY61473.1 GL453369 EFN76112.1 KK107293 EZA53364.1 LJIG01022701 KRT79162.1 CH933808 EDW08271.1 CH916367 EDW02391.1 GANO01003524 JAB56347.1 ADTU01001412 KQ435050 KZC14225.1 KQ435803 KOX73154.1 GEBQ01016266 JAT23711.1 QOIP01000005 RLU22773.1 CH480816 EDW47574.1 CM000362 CM002911 EDX06777.1 KMY93206.1 GBXI01010120 JAD04172.1 JXJN01026247 CCAG010005869 CH954179 EDV56308.1 AE013599 KX531367 AAM68685.3 ANY27177.1 CH963850 EDW74722.1 KQ414725 KOC62675.1 GECZ01027844 JAS41925.1 BT022606 AAY55022.1 GECU01000925 JAT06782.1 KQ981264 KYN43958.1 CH940667 EDW57141.1 BT081969 ACO53084.1 CM000158 EDW90761.1 CH902619 EDV35489.1 KK852692 KDR18339.1 CP012524 ALC41449.1 JTDY01000232 KOB78159.1 BT022693 AAY55109.1 GDAI01001771 JAI15832.1 DS235459 EEB15901.1 UFQS01000550 UFQT01000550 SSX04823.1 SSX25186.1 AK417203 BAN20418.1 GBHO01024047 GBRD01013140 GDHC01001510 JAG19557.1 JAG52686.1 JAQ17119.1 GDHF01018919 GDHF01010599 JAI33395.1 JAI41715.1 KK112864 KFM58721.1 GDKW01003287 JAI53308.1 KK854712 PTY18114.1 GAKP01000918 JAC58034.1 JH432008 GFTR01002623 JAW13803.1 GBGD01003314 JAC85575.1 AJVK01036842 CH479183 EDW35601.1 MWRG01017951 PRD23353.1
KQ460855 KPJ11898.1 KQ459601 KPI93986.1 BABH01004810 GALX01000386 JAB68080.1 GAPW01005589 GEHC01001452 JAC08009.1 JAV46193.1 DQ440480 CH477847 ABF18513.1 EAT35676.1 AXCN02000910 GEZM01070907 JAV66227.1 GGFJ01010391 MBW59532.1 AAAB01008900 EAA09360.5 APCN01002921 GAMD01002238 JAA99352.1 AXCM01001928 KQ971343 EFA03470.1 GFDL01011945 JAV23100.1 DS231842 EDS35277.1 GGFM01005518 MBW26269.1 GFDF01010872 JAV03212.1 ATLV01021826 KE525324 KFB46891.1 GGFK01011016 MBW44337.1 ADMH02002078 ETN59464.1 GL438984 EFN67968.1 AAZX01000344 NNAY01002545 OXU21090.1 KZ288215 PBC32779.1 JR051309 AEY61473.1 GL453369 EFN76112.1 KK107293 EZA53364.1 LJIG01022701 KRT79162.1 CH933808 EDW08271.1 CH916367 EDW02391.1 GANO01003524 JAB56347.1 ADTU01001412 KQ435050 KZC14225.1 KQ435803 KOX73154.1 GEBQ01016266 JAT23711.1 QOIP01000005 RLU22773.1 CH480816 EDW47574.1 CM000362 CM002911 EDX06777.1 KMY93206.1 GBXI01010120 JAD04172.1 JXJN01026247 CCAG010005869 CH954179 EDV56308.1 AE013599 KX531367 AAM68685.3 ANY27177.1 CH963850 EDW74722.1 KQ414725 KOC62675.1 GECZ01027844 JAS41925.1 BT022606 AAY55022.1 GECU01000925 JAT06782.1 KQ981264 KYN43958.1 CH940667 EDW57141.1 BT081969 ACO53084.1 CM000158 EDW90761.1 CH902619 EDV35489.1 KK852692 KDR18339.1 CP012524 ALC41449.1 JTDY01000232 KOB78159.1 BT022693 AAY55109.1 GDAI01001771 JAI15832.1 DS235459 EEB15901.1 UFQS01000550 UFQT01000550 SSX04823.1 SSX25186.1 AK417203 BAN20418.1 GBHO01024047 GBRD01013140 GDHC01001510 JAG19557.1 JAG52686.1 JAQ17119.1 GDHF01018919 GDHF01010599 JAI33395.1 JAI41715.1 KK112864 KFM58721.1 GDKW01003287 JAI53308.1 KK854712 PTY18114.1 GAKP01000918 JAC58034.1 JH432008 GFTR01002623 JAW13803.1 GBGD01003314 JAC85575.1 AJVK01036842 CH479183 EDW35601.1 MWRG01017951 PRD23353.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000005204
UP000008820
UP000075884
+ More
UP000075886 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000075885 UP000075883 UP000075920 UP000076407 UP000069272 UP000075881 UP000076408 UP000007266 UP000002320 UP000075901 UP000030765 UP000075880 UP000000673 UP000000311 UP000002358 UP000215335 UP000005203 UP000242457 UP000008237 UP000075900 UP000053097 UP000009192 UP000192223 UP000001070 UP000005205 UP000076502 UP000053105 UP000015102 UP000279307 UP000001292 UP000000304 UP000092445 UP000092460 UP000092444 UP000008711 UP000000803 UP000007798 UP000053825 UP000078541 UP000192221 UP000008792 UP000002282 UP000007801 UP000027135 UP000092553 UP000037510 UP000009046 UP000092443 UP000078200 UP000054359 UP000095300 UP000095301 UP000079169 UP000092462 UP000008744
UP000075886 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000075885 UP000075883 UP000075920 UP000076407 UP000069272 UP000075881 UP000076408 UP000007266 UP000002320 UP000075901 UP000030765 UP000075880 UP000000673 UP000000311 UP000002358 UP000215335 UP000005203 UP000242457 UP000008237 UP000075900 UP000053097 UP000009192 UP000192223 UP000001070 UP000005205 UP000076502 UP000053105 UP000015102 UP000279307 UP000001292 UP000000304 UP000092445 UP000092460 UP000092444 UP000008711 UP000000803 UP000007798 UP000053825 UP000078541 UP000192221 UP000008792 UP000002282 UP000007801 UP000027135 UP000092553 UP000037510 UP000009046 UP000092443 UP000078200 UP000054359 UP000095300 UP000095301 UP000079169 UP000092462 UP000008744
Interpro
Gene 3D
ProteinModelPortal
A0A1E1VY87
A0A212F7Y0
A0A194R284
A0A194PKM3
H9J900
V5GD49
+ More
A0A023EGS6 Q1HQG4 A0A182N804 A0A182QW26 A0A1Y1L558 A0A2M4C2J1 A0A182UYD0 A0A182TJR8 A0A182LBD2 Q7PQ38 A0A182HP58 A0A182PHE5 T1E923 A0A182LVI4 A0A182WAB7 A0A182XJA4 A0A182FH93 A0A182K137 A0A182YNR7 D6WLC5 A0A1Q3F6A8 B0W5F7 A0A2M3ZCP7 A0A1L8D9U5 A0A182SK74 A0A084W9J7 A0A182ISP2 A0A2M4AU81 W5J9P1 E2AF13 K7J575 A0A232ERW0 A0A088A1C6 A0A2A3ENS3 V9IK05 E2C7C5 A0A182RTQ1 A0A026WEA8 A0A0T6AVM8 B4KR84 A0A1W4XBD6 B4J8V1 U5EGA6 A0A158N9R5 A0A154PQM8 A0A0N0BFD2 T1H6D5 A0A1B6LJ33 A0A3L8DRF0 B4HPJ0 B4QCW6 A0A0A1WZ89 A0A1A9ZB61 A0A1B0C5U5 A0A1B0GBQ7 B3NS06 A1Z903 B4MRI3 A0A0L7QVN5 A0A1B6EVC5 Q4V5Q0 A0A1B6K5P1 A0A195FU22 A0A1W4UHG3 B4MFH0 C1C540 B4P599 B3MEM5 A0A067R500 A0A0M3QUY0 A0A0L7LS04 Q4V5G3 A0A0K8TN79 E0VR95 A0A336KPA6 R4WCV6 A0A1A9YS55 A0A1A9VPQ0 A0A0A9XFQ8 A0A0K8V488 A0A087T0T2 A0A0P4VQ54 A0A2R7WEM3 A0A034WTQ7 A0A1I8PI29 A0A1I8MLV0 A0A1S4EBV3 A0A3Q0ISJ7 T1JB36 A0A224XZU1 A0A069DP02 A0A1B0DM28 B4GGI1 A0A2P6KAL2
A0A023EGS6 Q1HQG4 A0A182N804 A0A182QW26 A0A1Y1L558 A0A2M4C2J1 A0A182UYD0 A0A182TJR8 A0A182LBD2 Q7PQ38 A0A182HP58 A0A182PHE5 T1E923 A0A182LVI4 A0A182WAB7 A0A182XJA4 A0A182FH93 A0A182K137 A0A182YNR7 D6WLC5 A0A1Q3F6A8 B0W5F7 A0A2M3ZCP7 A0A1L8D9U5 A0A182SK74 A0A084W9J7 A0A182ISP2 A0A2M4AU81 W5J9P1 E2AF13 K7J575 A0A232ERW0 A0A088A1C6 A0A2A3ENS3 V9IK05 E2C7C5 A0A182RTQ1 A0A026WEA8 A0A0T6AVM8 B4KR84 A0A1W4XBD6 B4J8V1 U5EGA6 A0A158N9R5 A0A154PQM8 A0A0N0BFD2 T1H6D5 A0A1B6LJ33 A0A3L8DRF0 B4HPJ0 B4QCW6 A0A0A1WZ89 A0A1A9ZB61 A0A1B0C5U5 A0A1B0GBQ7 B3NS06 A1Z903 B4MRI3 A0A0L7QVN5 A0A1B6EVC5 Q4V5Q0 A0A1B6K5P1 A0A195FU22 A0A1W4UHG3 B4MFH0 C1C540 B4P599 B3MEM5 A0A067R500 A0A0M3QUY0 A0A0L7LS04 Q4V5G3 A0A0K8TN79 E0VR95 A0A336KPA6 R4WCV6 A0A1A9YS55 A0A1A9VPQ0 A0A0A9XFQ8 A0A0K8V488 A0A087T0T2 A0A0P4VQ54 A0A2R7WEM3 A0A034WTQ7 A0A1I8PI29 A0A1I8MLV0 A0A1S4EBV3 A0A3Q0ISJ7 T1JB36 A0A224XZU1 A0A069DP02 A0A1B0DM28 B4GGI1 A0A2P6KAL2
Ontologies
GO
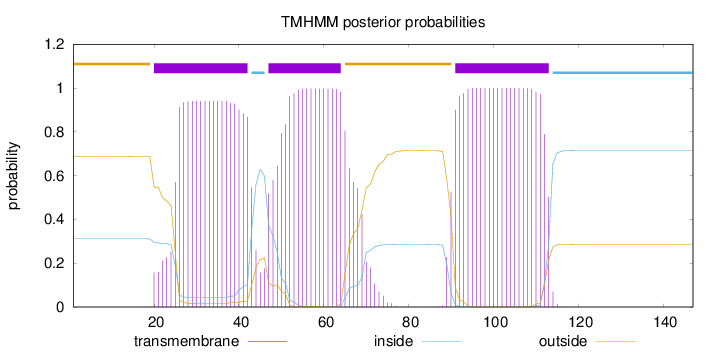
Topology
Length:
147
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
61.30004
Exp number, first 60 AAs:
30.76901
Total prob of N-in:
0.31204
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 46
TMhelix
47 - 64
outside
65 - 90
TMhelix
91 - 113
inside
114 - 147
Population Genetic Test Statistics
Pi
25.25225
Theta
185.710263
Tajima's D
-2.462644
CLR
0.712096
CSRT
0.000649967501624919
Interpretation
Uncertain
