Gene
KWMTBOMO01802 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006160
Annotation
L-threonine_dehydrogenase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.11 Peroxisomal Reliability : 1.455
Sequence
CDS
ATGAATAAATCCGCGAATATTCGACCCGATTGTGGCCTCGTTAACCTTGAAACTGACATTGAAATTTGTATAAGAAATATATGTAGAGCTTCGATTAAATTTAATGTAAGATTTTATTCGGGCGAATTATTAAGGAGAAGACCTCCGAAAATTTTGATTACCGGAGGACTTGGCCAATTGGGTGTCGAGTGTGCTAAATATCTCCGTGGGAAATATGGCCAGAACAACGTCATACTGTCAGACATTATTAAACCAACCGAAGAAATCGTTAAGGACGGCCCTTACATTTTCGTCGATATACTTGACTTCAAAGGACTTCAGAAAATCGTAGTAGATCACAGAGTCGATTGGTTGATTCATTTCTCGGCCCTACTCAGCGCAATAGGAGAACAAAACGTACCTTTAGCCGTAAGGGTCAATATAGAAGGCATGCATAATGTCATCGAGCTAGCTAAACAATACAAACTTCGGATATTTGTTCCTAGTACGATTGGAGCGTTCGGTCCTGATTCGCCAAGAAATCCAACGCCAAATATAACAATCCAAAGGCCGAGAACAATATACGGGGTGTCGAAAGTTCACGCGGAGTTGTTAGGAGAGTACTATTATCATAAATTCGGACTAGACTTCCGCTGTCTAAGATTCCCGGGAGTAATATCCAGTGATCCGCCAGGTGGAGGCACTACAGATTATGCAATCGCAATCTTCCATGACATTCTCCGTAGAGGAAATTACGAGTGTTATCTAAAACCAGACACGCGGCTTCCCATGATGCACGTTAGGGATGCCCTTCGAGCCCTATCTGAATACCTTGAGGCCCCAATTGAAACACTGAACAGACGTGTTTATAACGTTACTTCAATGAGCTTCACGCCGGAAGAATTGGTAGAAGCCATCAGCAAATATTTACCAGACTTGAAGATTACTTATAAGCCGGATAGTCGGCAAGATATAGCTGACTGGTGGCCGCAAGTGTTCGATGACAGCGAAGCTCGACGTGACTGGAACTGGAAACCTGAAGTTGACCTCGACAACTTGGTCAAGCTCATGCTGAAGGAAGTCAAAGAAAAAATAGCTGCGAATGGTCTTTGA
Protein
MNKSANIRPDCGLVNLETDIEICIRNICRASIKFNVRFYSGELLRRRPPKILITGGLGQLGVECAKYLRGKYGQNNVILSDIIKPTEEIVKDGPYIFVDILDFKGLQKIVVDHRVDWLIHFSALLSAIGEQNVPLAVRVNIEGMHNVIELAKQYKLRIFVPSTIGAFGPDSPRNPTPNITIQRPRTIYGVSKVHAELLGEYYYHKFGLDFRCLRFPGVISSDPPGGGTTDYAIAIFHDILRRGNYECYLKPDTRLPMMHVRDALRALSEYLEAPIETLNRRVYNVTSMSFTPEELVEAISKYLPDLKITYKPDSRQDIADWWPQVFDDSEARRDWNWKPEVDLDNLVKLMLKEVKEKIAANGL
Summary
Uniprot
Q3HR36
A0A2A4KAS4
H9J9G7
A0A194PKY0
A0A194R2H3
A0A2H1WKP5
+ More
A0A212EQA9 A0A212F7Y3 A0A0L0BNE5 B3M9A6 T1PKD7 A0A1A9XC88 A0A1B0B5A2 T1PG36 D3TLZ9 A0A034VMA3 A0A0M5IZ85 B4N6M0 A0A0A1X7A3 A0A182Y9J7 Q29DC8 B4H7M8 B4LGC5 B4KY45 B4IYZ4 A0A1W4VUK5 A0A3B0JTY1 A0A1I8NNT2 W8BFH5 A0A2M3ZF93 A0A182PC05 A0A182FB91 A0A2M4BRU2 A0A0K8WG63 B3NIM8 A0A2M4BSJ0 T1E7V5 A0A2M4BRU5 Q9VPE8 B4IUG8 B4IAB6 A0A1Y9IVY7 B4PET8 A0A182MDR4 A0A1S4F4P4 W5JTK1 Q7PZJ8 A0A182HHN3 Q17FE0 A0A182RXU6 A0A2M4A3E9 B4QS02 A0A182XJP9 A0A182URY1 A0A0L7LRU5 A0A1J1J9Q2 B0WLP8 A0A2M4A3N6 A0A2M4A438 A0A182N2R1 A0A182L580 A0A182QEG9 A0A182TBU0 A0A182JHM2 A0A084WIF7 A0A336LTD8 A0A2M4CM33 K7INW1 A0A1W4W754 A0A182KEP8 A0A182TDV1 A0A2J7PEE2 A0A026W624 A0A195B4J8 E2A9P7 A0A1L8E051 A0A158N8V6 E2C969 A0A195FP77 A0A195CU23 A0A232FM57 A0A2A3E8D3 A0A088AGS7 A0A151JCE7 A0A154P5C2 A0A0J7NHX9 A0A151WLC2 F4WML5 A0A0C9RD47 E0VPE4 A0A067RS69 A0A182H3Z0 A0A0L7R6I9 A0A0P4WK77 A0A1B0CH38 A0A0P5WUN5 A0A0N8AMW6 A0A0P5BV34 A0A0P6HUH1 A0A224Z7S4
A0A212EQA9 A0A212F7Y3 A0A0L0BNE5 B3M9A6 T1PKD7 A0A1A9XC88 A0A1B0B5A2 T1PG36 D3TLZ9 A0A034VMA3 A0A0M5IZ85 B4N6M0 A0A0A1X7A3 A0A182Y9J7 Q29DC8 B4H7M8 B4LGC5 B4KY45 B4IYZ4 A0A1W4VUK5 A0A3B0JTY1 A0A1I8NNT2 W8BFH5 A0A2M3ZF93 A0A182PC05 A0A182FB91 A0A2M4BRU2 A0A0K8WG63 B3NIM8 A0A2M4BSJ0 T1E7V5 A0A2M4BRU5 Q9VPE8 B4IUG8 B4IAB6 A0A1Y9IVY7 B4PET8 A0A182MDR4 A0A1S4F4P4 W5JTK1 Q7PZJ8 A0A182HHN3 Q17FE0 A0A182RXU6 A0A2M4A3E9 B4QS02 A0A182XJP9 A0A182URY1 A0A0L7LRU5 A0A1J1J9Q2 B0WLP8 A0A2M4A3N6 A0A2M4A438 A0A182N2R1 A0A182L580 A0A182QEG9 A0A182TBU0 A0A182JHM2 A0A084WIF7 A0A336LTD8 A0A2M4CM33 K7INW1 A0A1W4W754 A0A182KEP8 A0A182TDV1 A0A2J7PEE2 A0A026W624 A0A195B4J8 E2A9P7 A0A1L8E051 A0A158N8V6 E2C969 A0A195FP77 A0A195CU23 A0A232FM57 A0A2A3E8D3 A0A088AGS7 A0A151JCE7 A0A154P5C2 A0A0J7NHX9 A0A151WLC2 F4WML5 A0A0C9RD47 E0VPE4 A0A067RS69 A0A182H3Z0 A0A0L7R6I9 A0A0P4WK77 A0A1B0CH38 A0A0P5WUN5 A0A0N8AMW6 A0A0P5BV34 A0A0P6HUH1 A0A224Z7S4
Pubmed
19121390
26354079
22118469
26108605
17994087
25315136
+ More
20353571 25348373 25830018 25244985 15632085 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20920257 23761445 12364791 14747013 17210077 17510324 22936249 26227816 20966253 24438588 20075255 24508170 30249741 20798317 21347285 28648823 21719571 20566863 24845553 26483478 28797301
20353571 25348373 25830018 25244985 15632085 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20920257 23761445 12364791 14747013 17210077 17510324 22936249 26227816 20966253 24438588 20075255 24508170 30249741 20798317 21347285 28648823 21719571 20566863 24845553 26483478 28797301
EMBL
DQ202317
ABA43639.1
NWSH01000013
PCG80840.1
BABH01004809
BABH01004810
+ More
KQ459601 KPI93987.1 KQ460855 KPJ11897.1 ODYU01009320 SOQ53655.1 AGBW02013271 OWR43680.1 AGBW02009822 OWR49847.1 JRES01001603 KNC21552.1 CH902618 EDV40090.1 KA649164 AFP63793.1 JXJN01008650 KA646848 AFP61477.1 CCAG010023097 EZ422451 ADD18727.1 GAKP01016012 GAKP01016010 GAKP01016008 GAKP01016006 JAC42940.1 CP012525 ALC45111.1 CH964161 EDW80009.1 GBXI01007541 JAD06751.1 CH379070 EAL30486.2 CH479219 EDW34668.1 CH940647 EDW70454.1 CH933809 EDW18747.1 CH916366 EDV97702.1 OUUW01000002 SPP77149.1 GAMC01006490 JAC00066.1 GGFM01006473 MBW27224.1 GGFJ01006638 MBW55779.1 GDHF01002163 JAI50151.1 CH954178 EDV52524.1 GGFJ01006888 MBW56029.1 GAMD01002722 JAA98868.1 GGFJ01006639 MBW55780.1 AE014296 AY058410 AAF51607.1 AAL13639.1 CH891905 EDX00032.1 CH480826 EDW44229.1 CM000159 EDW95162.1 AXCM01006279 ADMH02000120 ETN67697.1 AAAB01008986 EAA00249.3 APCN01002431 CH477271 EAT45305.1 GGFK01002025 MBW35346.1 CM000363 CM002912 EDX11248.1 KMZ00805.1 JTDY01000232 KOB78152.1 CVRI01000073 CRL07745.1 DS231989 EDS30613.1 GGFK01002086 MBW35407.1 GGFK01002087 MBW35408.1 AXCN02001086 ATLV01023930 KE525347 KFB50001.1 UFQS01000099 UFQT01000099 SSW99499.1 SSX19879.1 GGFL01002224 MBW66402.1 NEVH01026098 PNF14711.1 KK107455 QOIP01000010 EZA50489.1 RLU17392.1 KQ976618 KYM79200.1 GL437918 EFN69849.1 GFDF01002047 JAV12037.1 ADTU01008821 ADTU01008822 GL453780 EFN75542.1 KQ981382 KYN42216.1 KQ977279 KYN04188.1 NNAY01000039 OXU31663.1 KZ288326 PBC27975.1 KQ979054 KYN23198.1 KQ434822 KZC07087.1 LBMM01004780 KMQ92090.1 KQ982959 KYQ48658.1 GL888218 EGI64492.1 GBYB01004836 JAG74603.1 DS235363 EEB15250.1 KK852498 KDR22649.1 JXUM01108452 JXUM01108453 JXUM01108454 KQ565232 KXJ71203.1 KQ414646 KOC66507.1 GDRN01031226 JAI67523.1 AJWK01011987 AJWK01011988 GDIP01081452 JAM22263.1 GDIQ01260131 LRGB01002167 JAJ91593.1 KZS08860.1 GDIP01179547 JAJ43855.1 GDIP01245858 GDIQ01013313 JAI77543.1 JAN81424.1 GFPF01012345 MAA23491.1
KQ459601 KPI93987.1 KQ460855 KPJ11897.1 ODYU01009320 SOQ53655.1 AGBW02013271 OWR43680.1 AGBW02009822 OWR49847.1 JRES01001603 KNC21552.1 CH902618 EDV40090.1 KA649164 AFP63793.1 JXJN01008650 KA646848 AFP61477.1 CCAG010023097 EZ422451 ADD18727.1 GAKP01016012 GAKP01016010 GAKP01016008 GAKP01016006 JAC42940.1 CP012525 ALC45111.1 CH964161 EDW80009.1 GBXI01007541 JAD06751.1 CH379070 EAL30486.2 CH479219 EDW34668.1 CH940647 EDW70454.1 CH933809 EDW18747.1 CH916366 EDV97702.1 OUUW01000002 SPP77149.1 GAMC01006490 JAC00066.1 GGFM01006473 MBW27224.1 GGFJ01006638 MBW55779.1 GDHF01002163 JAI50151.1 CH954178 EDV52524.1 GGFJ01006888 MBW56029.1 GAMD01002722 JAA98868.1 GGFJ01006639 MBW55780.1 AE014296 AY058410 AAF51607.1 AAL13639.1 CH891905 EDX00032.1 CH480826 EDW44229.1 CM000159 EDW95162.1 AXCM01006279 ADMH02000120 ETN67697.1 AAAB01008986 EAA00249.3 APCN01002431 CH477271 EAT45305.1 GGFK01002025 MBW35346.1 CM000363 CM002912 EDX11248.1 KMZ00805.1 JTDY01000232 KOB78152.1 CVRI01000073 CRL07745.1 DS231989 EDS30613.1 GGFK01002086 MBW35407.1 GGFK01002087 MBW35408.1 AXCN02001086 ATLV01023930 KE525347 KFB50001.1 UFQS01000099 UFQT01000099 SSW99499.1 SSX19879.1 GGFL01002224 MBW66402.1 NEVH01026098 PNF14711.1 KK107455 QOIP01000010 EZA50489.1 RLU17392.1 KQ976618 KYM79200.1 GL437918 EFN69849.1 GFDF01002047 JAV12037.1 ADTU01008821 ADTU01008822 GL453780 EFN75542.1 KQ981382 KYN42216.1 KQ977279 KYN04188.1 NNAY01000039 OXU31663.1 KZ288326 PBC27975.1 KQ979054 KYN23198.1 KQ434822 KZC07087.1 LBMM01004780 KMQ92090.1 KQ982959 KYQ48658.1 GL888218 EGI64492.1 GBYB01004836 JAG74603.1 DS235363 EEB15250.1 KK852498 KDR22649.1 JXUM01108452 JXUM01108453 JXUM01108454 KQ565232 KXJ71203.1 KQ414646 KOC66507.1 GDRN01031226 JAI67523.1 AJWK01011987 AJWK01011988 GDIP01081452 JAM22263.1 GDIQ01260131 LRGB01002167 JAJ91593.1 KZS08860.1 GDIP01179547 JAJ43855.1 GDIP01245858 GDIQ01013313 JAI77543.1 JAN81424.1 GFPF01012345 MAA23491.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000053240
UP000007151
UP000037069
+ More
UP000007801 UP000095301 UP000092443 UP000092460 UP000092444 UP000092553 UP000007798 UP000076408 UP000001819 UP000008744 UP000008792 UP000009192 UP000001070 UP000192221 UP000268350 UP000095300 UP000075885 UP000069272 UP000008711 UP000000803 UP000002282 UP000001292 UP000075920 UP000075883 UP000000673 UP000007062 UP000075840 UP000008820 UP000075900 UP000000304 UP000076407 UP000075903 UP000037510 UP000183832 UP000002320 UP000075884 UP000075882 UP000075886 UP000075901 UP000075880 UP000030765 UP000002358 UP000075881 UP000075902 UP000235965 UP000053097 UP000279307 UP000078540 UP000000311 UP000005205 UP000008237 UP000078541 UP000078542 UP000215335 UP000242457 UP000005203 UP000078492 UP000076502 UP000036403 UP000075809 UP000007755 UP000009046 UP000027135 UP000069940 UP000249989 UP000053825 UP000092461 UP000076858
UP000007801 UP000095301 UP000092443 UP000092460 UP000092444 UP000092553 UP000007798 UP000076408 UP000001819 UP000008744 UP000008792 UP000009192 UP000001070 UP000192221 UP000268350 UP000095300 UP000075885 UP000069272 UP000008711 UP000000803 UP000002282 UP000001292 UP000075920 UP000075883 UP000000673 UP000007062 UP000075840 UP000008820 UP000075900 UP000000304 UP000076407 UP000075903 UP000037510 UP000183832 UP000002320 UP000075884 UP000075882 UP000075886 UP000075901 UP000075880 UP000030765 UP000002358 UP000075881 UP000075902 UP000235965 UP000053097 UP000279307 UP000078540 UP000000311 UP000005205 UP000008237 UP000078541 UP000078542 UP000215335 UP000242457 UP000005203 UP000078492 UP000076502 UP000036403 UP000075809 UP000007755 UP000009046 UP000027135 UP000069940 UP000249989 UP000053825 UP000092461 UP000076858
Pfam
PF01370 Epimerase
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
Q3HR36
A0A2A4KAS4
H9J9G7
A0A194PKY0
A0A194R2H3
A0A2H1WKP5
+ More
A0A212EQA9 A0A212F7Y3 A0A0L0BNE5 B3M9A6 T1PKD7 A0A1A9XC88 A0A1B0B5A2 T1PG36 D3TLZ9 A0A034VMA3 A0A0M5IZ85 B4N6M0 A0A0A1X7A3 A0A182Y9J7 Q29DC8 B4H7M8 B4LGC5 B4KY45 B4IYZ4 A0A1W4VUK5 A0A3B0JTY1 A0A1I8NNT2 W8BFH5 A0A2M3ZF93 A0A182PC05 A0A182FB91 A0A2M4BRU2 A0A0K8WG63 B3NIM8 A0A2M4BSJ0 T1E7V5 A0A2M4BRU5 Q9VPE8 B4IUG8 B4IAB6 A0A1Y9IVY7 B4PET8 A0A182MDR4 A0A1S4F4P4 W5JTK1 Q7PZJ8 A0A182HHN3 Q17FE0 A0A182RXU6 A0A2M4A3E9 B4QS02 A0A182XJP9 A0A182URY1 A0A0L7LRU5 A0A1J1J9Q2 B0WLP8 A0A2M4A3N6 A0A2M4A438 A0A182N2R1 A0A182L580 A0A182QEG9 A0A182TBU0 A0A182JHM2 A0A084WIF7 A0A336LTD8 A0A2M4CM33 K7INW1 A0A1W4W754 A0A182KEP8 A0A182TDV1 A0A2J7PEE2 A0A026W624 A0A195B4J8 E2A9P7 A0A1L8E051 A0A158N8V6 E2C969 A0A195FP77 A0A195CU23 A0A232FM57 A0A2A3E8D3 A0A088AGS7 A0A151JCE7 A0A154P5C2 A0A0J7NHX9 A0A151WLC2 F4WML5 A0A0C9RD47 E0VPE4 A0A067RS69 A0A182H3Z0 A0A0L7R6I9 A0A0P4WK77 A0A1B0CH38 A0A0P5WUN5 A0A0N8AMW6 A0A0P5BV34 A0A0P6HUH1 A0A224Z7S4
A0A212EQA9 A0A212F7Y3 A0A0L0BNE5 B3M9A6 T1PKD7 A0A1A9XC88 A0A1B0B5A2 T1PG36 D3TLZ9 A0A034VMA3 A0A0M5IZ85 B4N6M0 A0A0A1X7A3 A0A182Y9J7 Q29DC8 B4H7M8 B4LGC5 B4KY45 B4IYZ4 A0A1W4VUK5 A0A3B0JTY1 A0A1I8NNT2 W8BFH5 A0A2M3ZF93 A0A182PC05 A0A182FB91 A0A2M4BRU2 A0A0K8WG63 B3NIM8 A0A2M4BSJ0 T1E7V5 A0A2M4BRU5 Q9VPE8 B4IUG8 B4IAB6 A0A1Y9IVY7 B4PET8 A0A182MDR4 A0A1S4F4P4 W5JTK1 Q7PZJ8 A0A182HHN3 Q17FE0 A0A182RXU6 A0A2M4A3E9 B4QS02 A0A182XJP9 A0A182URY1 A0A0L7LRU5 A0A1J1J9Q2 B0WLP8 A0A2M4A3N6 A0A2M4A438 A0A182N2R1 A0A182L580 A0A182QEG9 A0A182TBU0 A0A182JHM2 A0A084WIF7 A0A336LTD8 A0A2M4CM33 K7INW1 A0A1W4W754 A0A182KEP8 A0A182TDV1 A0A2J7PEE2 A0A026W624 A0A195B4J8 E2A9P7 A0A1L8E051 A0A158N8V6 E2C969 A0A195FP77 A0A195CU23 A0A232FM57 A0A2A3E8D3 A0A088AGS7 A0A151JCE7 A0A154P5C2 A0A0J7NHX9 A0A151WLC2 F4WML5 A0A0C9RD47 E0VPE4 A0A067RS69 A0A182H3Z0 A0A0L7R6I9 A0A0P4WK77 A0A1B0CH38 A0A0P5WUN5 A0A0N8AMW6 A0A0P5BV34 A0A0P6HUH1 A0A224Z7S4
PDB
4YRA
E-value=1.82686e-107,
Score=994
Ontologies
PATHWAY
GO
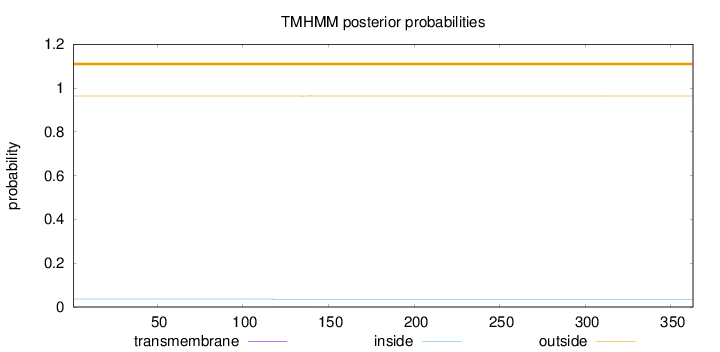
Topology
Length:
363
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02379
Exp number, first 60 AAs:
0.00175
Total prob of N-in:
0.03685
outside
1 - 363
Population Genetic Test Statistics
Pi
32.399769
Theta
207.877639
Tajima's D
0.555213
CLR
0.415481
CSRT
0.529173541322934
Interpretation
Uncertain
