Gene
KWMTBOMO01797
Pre Gene Modal
BGIBMGA005996
Annotation
PREDICTED:_inactive_peptidyl-prolyl_cis-trans_isomerase_shutdown-like_[Papilio_xuthus]
Full name
Peptidylprolyl isomerase
+ More
Inactive peptidyl-prolyl cis-trans isomerase shutdown
Inactive peptidyl-prolyl cis-trans isomerase shutdown
Location in the cell
Extracellular Reliability : 1.58 Mitochondrial Reliability : 1.108
Sequence
CDS
ATGTGTACCGTCACTTTAGAAAATGGACTAGATCTACGGGAATGTACAACTACAGGCTCTATTTTGCACATCAATGAGATATATGAAGAAAATGATGATAATGATACTACAGAGAGCAAAGTATTTAAGACAATTGACATTCTGGGAGAACCTGTAAAAAATTTTGAAGTCTTAAAAAATGAATTGATTCCTGTTGATGAAAATCACTATGTAATTAAGAAAATACTAGAAACTGGTGGAGGTATGCCCTTGCATGATGGATGCACAGTCTCAATAGCATTCTCTGGATACTGGGAAAATGAGTTACAACCATTTGACGTAATGAGTTTAAATAATCCAATGACTGTAGATCTCAAAGACAGTGGTTTACTACCAGGACTAGATATAGCTGTTAGATCAATGCTTGTTGTGCACTGA
Protein
MCTVTLENGLDLRECTTTGSILHINEIYEENDDNDTTESKVFKTIDILGEPVKNFEVLKNELIPVDENHYVIKKILETGGGMPLHDGCTVSIAFSGYWENELQPFDVMSLNNPMTVDLKDSGLLPGLDIAVRSMLVVH
Summary
Description
Co-chaperone required during oogenesis to repress transposable elements and prevent their mobilization, which is essential for the germline integrity. Acts via the piRNA metabolic process, which mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and govern the methylation and subsequent repression of transposons. Acts as a co-chaperone via its interaction with Hsp83/HSP90 and is required for the biogenesis of all three piRNA major populations.
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Subunit
Interacts with Hsp83.
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Belongs to the FKBP6 family.
Belongs to the AB hydrolase superfamily. Lipase family.
Belongs to the FKBP6 family.
Belongs to the AB hydrolase superfamily. Lipase family.
Keywords
Complete proteome
Cytoplasm
Differentiation
Meiosis
Oogenesis
Reference proteome
Repeat
RNA-mediated gene silencing
TPR repeat
Feature
chain Inactive peptidyl-prolyl cis-trans isomerase shutdown
Uniprot
H9J904
A0A2A4KA46
A0A194PKM8
A0A194R279
A0A2H1X0J5
A0A212EQC1
+ More
S4PB25 A0A0L7K2F0 A0A0L7LRS1 A0A240PJX4 A0A2L2YF89 A0A182MGT8 A0A084VU23 A0A084VV77 T1IHK7 A0A182T071 A0A182YPF5 W5JRX6 A0A182Q1X1 A0A0P4W9F9 A0A2S2PM08 A0A182FTD7 A0A182JWF1 A0A182WF59 A0A0M5IZM0 A0A1W4XBG3 A0A2H8TG04 A0A151IEV6 A0A182R8Q0 Q9W1I9 F6UPL4 A0A1W2WD62 T1IJ05 E2A4Y8 A0A2A3EFJ9 A0A2R7VTC0 A0A2T7PCS5 A0A131YQH4 A0A2R5L7K0 A0A2S2QU73 A0A2S2Q1C7 A0A3B0JKR6 B4QB02 B4I8U0 E2C0E2 J9K208 Q293G0 B4GBK4 A0A0J7L1A3 A0A0A1XP56 A0A2S2NAH8
S4PB25 A0A0L7K2F0 A0A0L7LRS1 A0A240PJX4 A0A2L2YF89 A0A182MGT8 A0A084VU23 A0A084VV77 T1IHK7 A0A182T071 A0A182YPF5 W5JRX6 A0A182Q1X1 A0A0P4W9F9 A0A2S2PM08 A0A182FTD7 A0A182JWF1 A0A182WF59 A0A0M5IZM0 A0A1W4XBG3 A0A2H8TG04 A0A151IEV6 A0A182R8Q0 Q9W1I9 F6UPL4 A0A1W2WD62 T1IJ05 E2A4Y8 A0A2A3EFJ9 A0A2R7VTC0 A0A2T7PCS5 A0A131YQH4 A0A2R5L7K0 A0A2S2QU73 A0A2S2Q1C7 A0A3B0JKR6 B4QB02 B4I8U0 E2C0E2 J9K208 Q293G0 B4GBK4 A0A0J7L1A3 A0A0A1XP56 A0A2S2NAH8
EC Number
5.2.1.8
Pubmed
EMBL
BABH01004803
BABH01004804
NWSH01000013
PCG80856.1
KQ459601
KPI93991.1
+ More
KQ460855 KPJ11893.1 ODYU01012494 SOQ58831.1 AGBW02013271 OWR43685.1 GAIX01004776 JAA87784.1 JTDY01017280 KOB51858.1 JTDY01000232 KOB78158.1 IAAA01022285 IAAA01022286 LAA06687.1 AXCM01000254 ATLV01016628 KE525098 KFB41467.1 ATLV01017157 KE525157 KFB41871.1 JH429910 ADMH02000540 ETN66048.1 AXCN02002114 GDRN01070793 GDRN01070791 JAI63809.1 GGMR01017297 MBY29916.1 CP012524 ALC40886.1 GFXV01001194 MBW12999.1 KQ977854 KYM99352.1 AE013599 AY058554 EAAA01001361 JH430221 GL436770 EFN71505.1 KZ288269 PBC29989.1 KK854074 PTY10763.1 PZQS01000004 PVD31214.1 GEDV01007053 JAP81504.1 GGLE01001345 MBY05471.1 GGMS01011877 MBY81080.1 GGMS01002365 MBY71568.1 OUUW01000001 SPP75950.1 CM000362 CM002911 EDX08439.1 KMY96126.1 CH480824 EDW57015.1 GL451770 EFN78601.1 ABLF02022443 ABLF02022445 ABLF02022446 ABLF02061685 CM000071 EAL24651.2 CH479181 EDW31299.1 LBMM01001374 KMQ96426.1 GBXI01001622 JAD12670.1 GGMR01001167 MBY13786.1
KQ460855 KPJ11893.1 ODYU01012494 SOQ58831.1 AGBW02013271 OWR43685.1 GAIX01004776 JAA87784.1 JTDY01017280 KOB51858.1 JTDY01000232 KOB78158.1 IAAA01022285 IAAA01022286 LAA06687.1 AXCM01000254 ATLV01016628 KE525098 KFB41467.1 ATLV01017157 KE525157 KFB41871.1 JH429910 ADMH02000540 ETN66048.1 AXCN02002114 GDRN01070793 GDRN01070791 JAI63809.1 GGMR01017297 MBY29916.1 CP012524 ALC40886.1 GFXV01001194 MBW12999.1 KQ977854 KYM99352.1 AE013599 AY058554 EAAA01001361 JH430221 GL436770 EFN71505.1 KZ288269 PBC29989.1 KK854074 PTY10763.1 PZQS01000004 PVD31214.1 GEDV01007053 JAP81504.1 GGLE01001345 MBY05471.1 GGMS01011877 MBY81080.1 GGMS01002365 MBY71568.1 OUUW01000001 SPP75950.1 CM000362 CM002911 EDX08439.1 KMY96126.1 CH480824 EDW57015.1 GL451770 EFN78601.1 ABLF02022443 ABLF02022445 ABLF02022446 ABLF02061685 CM000071 EAL24651.2 CH479181 EDW31299.1 LBMM01001374 KMQ96426.1 GBXI01001622 JAD12670.1 GGMR01001167 MBY13786.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000075880 UP000075883 UP000030765 UP000075901 UP000076408 UP000000673 UP000075886 UP000069272 UP000075881 UP000075920 UP000092553 UP000192223 UP000078542 UP000075900 UP000000803 UP000008144 UP000000311 UP000242457 UP000245119 UP000268350 UP000000304 UP000001292 UP000008237 UP000007819 UP000001819 UP000008744 UP000036403
UP000075880 UP000075883 UP000030765 UP000075901 UP000076408 UP000000673 UP000075886 UP000069272 UP000075881 UP000075920 UP000092553 UP000192223 UP000078542 UP000075900 UP000000803 UP000008144 UP000000311 UP000242457 UP000245119 UP000268350 UP000000304 UP000001292 UP000008237 UP000007819 UP000001819 UP000008744 UP000036403
Pfam
Interpro
IPR013026
TPR-contain_dom
+ More
IPR011990 TPR-like_helical_dom_sf
IPR001179 PPIase_FKBP_dom
IPR042282 FKBP6/shu
IPR019734 TPR_repeat
IPR036028 SH3-like_dom_sf
IPR041794 MyoVII_FERM_C2
IPR027417 P-loop_NTPase
IPR000299 FERM_domain
IPR029071 Ubiquitin-like_domsf
IPR018979 FERM_N
IPR036961 Kinesin_motor_dom_sf
IPR019748 FERM_central
IPR001609 Myosin_head_motor_dom
IPR011993 PH-like_dom_sf
IPR041793 MyoVII_FERM_C1
IPR038185 MyTH4_dom_sf
IPR035963 FERM_2
IPR036106 MYSc_Myo7
IPR000048 IQ_motif_EF-hand-BS
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR019749 Band_41_domain
IPR001452 SH3_domain
IPR000857 MyTH4_dom
IPR018490 cNMP-bd-like
IPR014710 RmlC-like_jellyroll
IPR018488 cNMP-bd_CS
IPR000595 cNMP-bd_dom
IPR005821 Ion_trans_dom
IPR013105 TPR_2
IPR036867 R3H_dom_sf
IPR007277 Svp26/Tex261
IPR023566 PPIase_FKBP
IPR000734 TAG_lipase
IPR029058 AB_hydrolase
IPR004117 7tm6_olfct_rcpt
IPR013818 Lipase/vitellogenin
IPR033906 Lipase_N
IPR011990 TPR-like_helical_dom_sf
IPR001179 PPIase_FKBP_dom
IPR042282 FKBP6/shu
IPR019734 TPR_repeat
IPR036028 SH3-like_dom_sf
IPR041794 MyoVII_FERM_C2
IPR027417 P-loop_NTPase
IPR000299 FERM_domain
IPR029071 Ubiquitin-like_domsf
IPR018979 FERM_N
IPR036961 Kinesin_motor_dom_sf
IPR019748 FERM_central
IPR001609 Myosin_head_motor_dom
IPR011993 PH-like_dom_sf
IPR041793 MyoVII_FERM_C1
IPR038185 MyTH4_dom_sf
IPR035963 FERM_2
IPR036106 MYSc_Myo7
IPR000048 IQ_motif_EF-hand-BS
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR019749 Band_41_domain
IPR001452 SH3_domain
IPR000857 MyTH4_dom
IPR018490 cNMP-bd-like
IPR014710 RmlC-like_jellyroll
IPR018488 cNMP-bd_CS
IPR000595 cNMP-bd_dom
IPR005821 Ion_trans_dom
IPR013105 TPR_2
IPR036867 R3H_dom_sf
IPR007277 Svp26/Tex261
IPR023566 PPIase_FKBP
IPR000734 TAG_lipase
IPR029058 AB_hydrolase
IPR004117 7tm6_olfct_rcpt
IPR013818 Lipase/vitellogenin
IPR033906 Lipase_N
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J904
A0A2A4KA46
A0A194PKM8
A0A194R279
A0A2H1X0J5
A0A212EQC1
+ More
S4PB25 A0A0L7K2F0 A0A0L7LRS1 A0A240PJX4 A0A2L2YF89 A0A182MGT8 A0A084VU23 A0A084VV77 T1IHK7 A0A182T071 A0A182YPF5 W5JRX6 A0A182Q1X1 A0A0P4W9F9 A0A2S2PM08 A0A182FTD7 A0A182JWF1 A0A182WF59 A0A0M5IZM0 A0A1W4XBG3 A0A2H8TG04 A0A151IEV6 A0A182R8Q0 Q9W1I9 F6UPL4 A0A1W2WD62 T1IJ05 E2A4Y8 A0A2A3EFJ9 A0A2R7VTC0 A0A2T7PCS5 A0A131YQH4 A0A2R5L7K0 A0A2S2QU73 A0A2S2Q1C7 A0A3B0JKR6 B4QB02 B4I8U0 E2C0E2 J9K208 Q293G0 B4GBK4 A0A0J7L1A3 A0A0A1XP56 A0A2S2NAH8
S4PB25 A0A0L7K2F0 A0A0L7LRS1 A0A240PJX4 A0A2L2YF89 A0A182MGT8 A0A084VU23 A0A084VV77 T1IHK7 A0A182T071 A0A182YPF5 W5JRX6 A0A182Q1X1 A0A0P4W9F9 A0A2S2PM08 A0A182FTD7 A0A182JWF1 A0A182WF59 A0A0M5IZM0 A0A1W4XBG3 A0A2H8TG04 A0A151IEV6 A0A182R8Q0 Q9W1I9 F6UPL4 A0A1W2WD62 T1IJ05 E2A4Y8 A0A2A3EFJ9 A0A2R7VTC0 A0A2T7PCS5 A0A131YQH4 A0A2R5L7K0 A0A2S2QU73 A0A2S2Q1C7 A0A3B0JKR6 B4QB02 B4I8U0 E2C0E2 J9K208 Q293G0 B4GBK4 A0A0J7L1A3 A0A0A1XP56 A0A2S2NAH8
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Secreted
Secreted
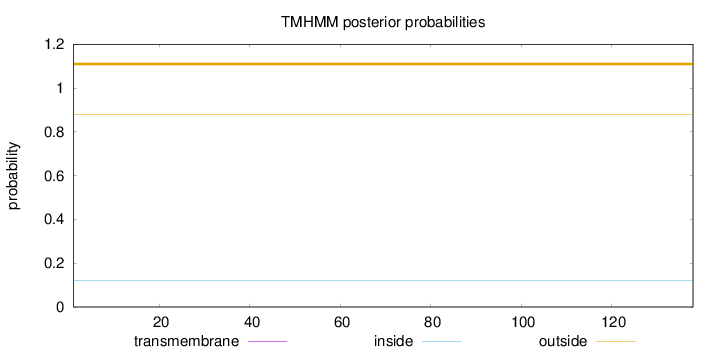
Length:
138
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00613999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12109
outside
1 - 138
Population Genetic Test Statistics
Pi
6.76472
Theta
36.982272
Tajima's D
-2.508192
CLR
342.905562
CSRT
0.000699965001749912
Interpretation
Possibly Positive selection
