Pre Gene Modal
BGIBMGA006154
Annotation
mannose-P-dolichol_utilization_defect_1_protein_[Bombyx_mori]
Full name
Mannose-P-dolichol utilization defect 1 protein homolog
Location in the cell
PlasmaMembrane Reliability : 4.894
Sequence
CDS
ATGGCTGAAATTTTAAAAGGATTATTATTAGGGGTGTTGTCTCAAAAGTGTTACAACGAATACTTTCTAAAGTACAATTTTCTTGATGTACCATGTTTCAAATCCACACTCAGCAAAGGTCTCGGAATCGGCATCATTCTAGGGTCCATACTGGTAAAAGTGCCACAAATTTTCAAAATTTTACAAAGTAAGAGTGCCGAAGGAATCAATATTTATGGAGTTTATCTGGAGTTGTTTGCTATCACTGCGAACTTTGCTTATAGCTATGTAATGGGTTTCCCATTCAGTGCATGGGGTGAAGGAACATTTTTAGCCATCCAAACGGCAATGATAGCAGCTTTAGTACTCCATTATGGTGGCGCTCCAATGAAGGGCGGAATATTTCTGTCTGTTTACTGTGCAATAGTCTCAGTTTTAGTAAGTGGTTATACCTCCACTGATATATTATGGACCATGCAGGCTGTCACTGTTCCTATCATTCTCATAGCCAAGTCAATCCAAATAGGAACGAACTACAAAAATGGCAGCACTGGTCAACTGTCGTTTATAACCTGCTTCCTGTTGTTTGGAGGTAGTGTAGCAAGAATCTTCACATCAATTCAAGAAACTGGAGACTCCATTATTATTCTGACCTACTGTGTTTCAACCATTGCCAATGGTGCTATTGTTTTACAAATGTTGTGGTACTGGAATGTTGAAAAGACAAAGTAA
Protein
MAEILKGLLLGVLSQKCYNEYFLKYNFLDVPCFKSTLSKGLGIGIILGSILVKVPQIFKILQSKSAEGINIYGVYLELFAITANFAYSYVMGFPFSAWGEGTFLAIQTAMIAALVLHYGGAPMKGGIFLSVYCAIVSVLVSGYTSTDILWTMQAVTVPIILIAKSIQIGTNYKNGSTGQLSFITCFLLFGGSVARIFTSIQETGDSIIILTYCVSTIANGAIVLQMLWYWNVEKTK
Summary
Similarity
Belongs to the MPDU1 (TC 2.A.43.3) family.
Keywords
Complete proteome
Membrane
Reference proteome
Repeat
Transmembrane
Transmembrane helix
Transport
Feature
chain Mannose-P-dolichol utilization defect 1 protein homolog
Uniprot
Q2F5W2
A0A194R2G3
I4DL06
A0A2A4KAM5
A0A194PKZ0
A0A212F7E3
+ More
A0A1E1W1Y2 A0A2H1WVY8 S4PYG5 A0A0L7KY04 A0A336LMD6 A0A0K8TTA9 A0A023ELJ1 A0A182H1B5 U5EZL0 B4LQK4 A0A158P471 A0A195CTT7 A0A026WJJ8 B4GSH5 A0A1L8DTX1 Q29L16 F4WIQ6 A0A1J1I2E6 T1PDY6 A0A1L8EIV5 A0A151JRC9 A0A1L8EIP6 A0A1I8NNK4 A0A195FPA4 A0A0M3QTR3 B3MP76 A0A1B0GHY6 A0A3B0JNK3 B4JR23 A0A1B6D008 B4KFM6 B3N4D4 V5GVL0 A0A1W4UVL7 B4N0K6 B4I1A2 B4NW91 A0A2A3ECP4 A0A088AGR8 Q9VMW8 A0A1W4X3F3 B4Q361 A0A1S3SEA5 A0A060WHI1 A0A0L0CDH3 A0A182J0R7 A0A182IC72 A0A1S3N8K7 A0A0C9R4Z4 A0A182L0K0 A0A182UNM9 A0A182U4R2 Q7Q4F6 A0A2M4CV78 A0A1S4GY33 A0A3P8SND2 A0A2M4AK58 A0A3P8ZUS6 A0A182XFY5 A0A182JT48 A0A3B4Z3C1 A0A182YDV0 A0A0J7L6B5 A0A3P8WC14 A0A0A1WTE1 W8C5V8 A0A084W5X9 A0A182PLQ8 A0A182NTZ1 A0A182W6G3 A0A1Y1NKW7 A0A0K8VIM1 T1DQF3 A0A226EPG8 A0A182RDQ7 A0A182QRD4 A0A0P4WIT3 A0A182M7S6 A0A3B4TK66 A0A034WP28 A0A151WLL4 A0A0P5YRT6 A0A3B4XH03 A0A3P9BSK3 A0A3B1K2H6 E9IIQ0 A0A0P4WN66 A0A1Q3FFI2 A0A154P598 A0A3P8PQ78 A0A0P5V1G1 E9H6P3 I3IVW7 A0A060YB50
A0A1E1W1Y2 A0A2H1WVY8 S4PYG5 A0A0L7KY04 A0A336LMD6 A0A0K8TTA9 A0A023ELJ1 A0A182H1B5 U5EZL0 B4LQK4 A0A158P471 A0A195CTT7 A0A026WJJ8 B4GSH5 A0A1L8DTX1 Q29L16 F4WIQ6 A0A1J1I2E6 T1PDY6 A0A1L8EIV5 A0A151JRC9 A0A1L8EIP6 A0A1I8NNK4 A0A195FPA4 A0A0M3QTR3 B3MP76 A0A1B0GHY6 A0A3B0JNK3 B4JR23 A0A1B6D008 B4KFM6 B3N4D4 V5GVL0 A0A1W4UVL7 B4N0K6 B4I1A2 B4NW91 A0A2A3ECP4 A0A088AGR8 Q9VMW8 A0A1W4X3F3 B4Q361 A0A1S3SEA5 A0A060WHI1 A0A0L0CDH3 A0A182J0R7 A0A182IC72 A0A1S3N8K7 A0A0C9R4Z4 A0A182L0K0 A0A182UNM9 A0A182U4R2 Q7Q4F6 A0A2M4CV78 A0A1S4GY33 A0A3P8SND2 A0A2M4AK58 A0A3P8ZUS6 A0A182XFY5 A0A182JT48 A0A3B4Z3C1 A0A182YDV0 A0A0J7L6B5 A0A3P8WC14 A0A0A1WTE1 W8C5V8 A0A084W5X9 A0A182PLQ8 A0A182NTZ1 A0A182W6G3 A0A1Y1NKW7 A0A0K8VIM1 T1DQF3 A0A226EPG8 A0A182RDQ7 A0A182QRD4 A0A0P4WIT3 A0A182M7S6 A0A3B4TK66 A0A034WP28 A0A151WLL4 A0A0P5YRT6 A0A3B4XH03 A0A3P9BSK3 A0A3B1K2H6 E9IIQ0 A0A0P4WN66 A0A1Q3FFI2 A0A154P598 A0A3P8PQ78 A0A0P5V1G1 E9H6P3 I3IVW7 A0A060YB50
Pubmed
19121390
26354079
22651552
22118469
23622113
26227816
+ More
26369729 24945155 26483478 17994087 21347285 24508170 30249741 15632085 21719571 25315136 17550304 10731132 12537572 22936249 24755649 26108605 20966253 12364791 25069045 25244985 24487278 25830018 24495485 24438588 28004739 25348373 25186727 25329095 21282665 21292972
26369729 24945155 26483478 17994087 21347285 24508170 30249741 15632085 21719571 25315136 17550304 10731132 12537572 22936249 24755649 26108605 20966253 12364791 25069045 25244985 24487278 25830018 24495485 24438588 28004739 25348373 25186727 25329095 21282665 21292972
EMBL
BABH01004776
DQ311311
ABD36255.1
KQ460855
KPJ11887.1
AK401974
+ More
BAM18596.1 NWSH01000013 PCG80843.1 KQ459601 KPI93997.1 AGBW02009882 OWR49652.1 GDQN01010068 JAT80986.1 ODYU01011427 SOQ57162.1 GAIX01003328 JAA89232.1 JTDY01004642 KOB67919.1 UFQT01000016 SSX17813.1 GDAI01000448 JAI17155.1 GAPW01003501 JAC10097.1 JXUM01102974 KQ564768 KXJ71768.1 GANO01000601 JAB59270.1 CH940649 EDW64461.1 ADTU01008677 KQ977279 KYN04108.1 KK107167 QOIP01000010 EZA56222.1 RLU17528.1 CH479189 EDW25334.1 GFDF01004247 JAV09837.1 CH379061 EAL33008.1 GL888177 EGI65852.1 CVRI01000038 CRK93938.1 KA646355 AFP60984.1 GFDG01000276 JAV18523.1 KQ978615 KYN29844.1 GFDG01000275 JAV18524.1 KQ981382 KYN42288.1 CP012523 ALC39339.1 CH902620 EDV32195.1 AJWK01010436 OUUW01000006 SPP81942.1 CH916372 EDV99353.1 GEDC01018268 GEDC01012757 JAS19030.1 JAS24541.1 CH933807 EDW13141.1 CH954177 EDV57804.1 GALX01004193 JAB64273.1 CH963920 EDW77619.1 CH480820 EDW54309.1 CM000157 EDW88408.1 KZ288283 PBC29515.1 AE014134 BT004850 CM000361 CM002910 EDX03777.1 KMY88192.1 FR904457 CDQ64010.1 JRES01000536 KNC30310.1 APCN01000778 GBYB01011379 JAG81146.1 AAAB01008964 EAA12192.3 GGFL01004560 MBW68738.1 GGFK01007855 MBW41176.1 LBMM01000547 KMQ98083.1 GBXI01011993 JAD02299.1 GAMC01004654 JAC01902.1 ATLV01020706 KE525305 KFB45623.1 GEZM01003516 JAV96756.1 GDHF01013585 JAI38729.1 GAMD01002544 JAA99046.1 LNIX01000002 OXA59098.1 AXCN02000807 GDRN01070185 JAI63923.1 AXCM01000782 GAKP01003007 JAC55945.1 KQ982959 KYQ48736.1 GDIP01054019 JAM49696.1 GL763505 EFZ19556.1 GDIP01253094 JAI70307.1 GFDL01008739 JAV26306.1 KQ434822 KZC07099.1 GDIP01108453 JAL95261.1 GL732598 EFX72543.1 AERX01054082 AERX01054083 FR908899 CDQ88717.1
BAM18596.1 NWSH01000013 PCG80843.1 KQ459601 KPI93997.1 AGBW02009882 OWR49652.1 GDQN01010068 JAT80986.1 ODYU01011427 SOQ57162.1 GAIX01003328 JAA89232.1 JTDY01004642 KOB67919.1 UFQT01000016 SSX17813.1 GDAI01000448 JAI17155.1 GAPW01003501 JAC10097.1 JXUM01102974 KQ564768 KXJ71768.1 GANO01000601 JAB59270.1 CH940649 EDW64461.1 ADTU01008677 KQ977279 KYN04108.1 KK107167 QOIP01000010 EZA56222.1 RLU17528.1 CH479189 EDW25334.1 GFDF01004247 JAV09837.1 CH379061 EAL33008.1 GL888177 EGI65852.1 CVRI01000038 CRK93938.1 KA646355 AFP60984.1 GFDG01000276 JAV18523.1 KQ978615 KYN29844.1 GFDG01000275 JAV18524.1 KQ981382 KYN42288.1 CP012523 ALC39339.1 CH902620 EDV32195.1 AJWK01010436 OUUW01000006 SPP81942.1 CH916372 EDV99353.1 GEDC01018268 GEDC01012757 JAS19030.1 JAS24541.1 CH933807 EDW13141.1 CH954177 EDV57804.1 GALX01004193 JAB64273.1 CH963920 EDW77619.1 CH480820 EDW54309.1 CM000157 EDW88408.1 KZ288283 PBC29515.1 AE014134 BT004850 CM000361 CM002910 EDX03777.1 KMY88192.1 FR904457 CDQ64010.1 JRES01000536 KNC30310.1 APCN01000778 GBYB01011379 JAG81146.1 AAAB01008964 EAA12192.3 GGFL01004560 MBW68738.1 GGFK01007855 MBW41176.1 LBMM01000547 KMQ98083.1 GBXI01011993 JAD02299.1 GAMC01004654 JAC01902.1 ATLV01020706 KE525305 KFB45623.1 GEZM01003516 JAV96756.1 GDHF01013585 JAI38729.1 GAMD01002544 JAA99046.1 LNIX01000002 OXA59098.1 AXCN02000807 GDRN01070185 JAI63923.1 AXCM01000782 GAKP01003007 JAC55945.1 KQ982959 KYQ48736.1 GDIP01054019 JAM49696.1 GL763505 EFZ19556.1 GDIP01253094 JAI70307.1 GFDL01008739 JAV26306.1 KQ434822 KZC07099.1 GDIP01108453 JAL95261.1 GL732598 EFX72543.1 AERX01054082 AERX01054083 FR908899 CDQ88717.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000037510
+ More
UP000069940 UP000249989 UP000008792 UP000005205 UP000078542 UP000053097 UP000279307 UP000008744 UP000001819 UP000007755 UP000183832 UP000095301 UP000078492 UP000095300 UP000078541 UP000092553 UP000007801 UP000092461 UP000268350 UP000001070 UP000009192 UP000008711 UP000192221 UP000007798 UP000001292 UP000002282 UP000242457 UP000005203 UP000000803 UP000192223 UP000000304 UP000087266 UP000193380 UP000037069 UP000075880 UP000075840 UP000075882 UP000075903 UP000075902 UP000007062 UP000265080 UP000265140 UP000076407 UP000075881 UP000261400 UP000076408 UP000036403 UP000265120 UP000030765 UP000075885 UP000075884 UP000075920 UP000198287 UP000075900 UP000075886 UP000075883 UP000261420 UP000075809 UP000261360 UP000265160 UP000018467 UP000076502 UP000265100 UP000000305 UP000005207
UP000069940 UP000249989 UP000008792 UP000005205 UP000078542 UP000053097 UP000279307 UP000008744 UP000001819 UP000007755 UP000183832 UP000095301 UP000078492 UP000095300 UP000078541 UP000092553 UP000007801 UP000092461 UP000268350 UP000001070 UP000009192 UP000008711 UP000192221 UP000007798 UP000001292 UP000002282 UP000242457 UP000005203 UP000000803 UP000192223 UP000000304 UP000087266 UP000193380 UP000037069 UP000075880 UP000075840 UP000075882 UP000075903 UP000075902 UP000007062 UP000265080 UP000265140 UP000076407 UP000075881 UP000261400 UP000076408 UP000036403 UP000265120 UP000030765 UP000075885 UP000075884 UP000075920 UP000198287 UP000075900 UP000075886 UP000075883 UP000261420 UP000075809 UP000261360 UP000265160 UP000018467 UP000076502 UP000265100 UP000000305 UP000005207
Interpro
ProteinModelPortal
Q2F5W2
A0A194R2G3
I4DL06
A0A2A4KAM5
A0A194PKZ0
A0A212F7E3
+ More
A0A1E1W1Y2 A0A2H1WVY8 S4PYG5 A0A0L7KY04 A0A336LMD6 A0A0K8TTA9 A0A023ELJ1 A0A182H1B5 U5EZL0 B4LQK4 A0A158P471 A0A195CTT7 A0A026WJJ8 B4GSH5 A0A1L8DTX1 Q29L16 F4WIQ6 A0A1J1I2E6 T1PDY6 A0A1L8EIV5 A0A151JRC9 A0A1L8EIP6 A0A1I8NNK4 A0A195FPA4 A0A0M3QTR3 B3MP76 A0A1B0GHY6 A0A3B0JNK3 B4JR23 A0A1B6D008 B4KFM6 B3N4D4 V5GVL0 A0A1W4UVL7 B4N0K6 B4I1A2 B4NW91 A0A2A3ECP4 A0A088AGR8 Q9VMW8 A0A1W4X3F3 B4Q361 A0A1S3SEA5 A0A060WHI1 A0A0L0CDH3 A0A182J0R7 A0A182IC72 A0A1S3N8K7 A0A0C9R4Z4 A0A182L0K0 A0A182UNM9 A0A182U4R2 Q7Q4F6 A0A2M4CV78 A0A1S4GY33 A0A3P8SND2 A0A2M4AK58 A0A3P8ZUS6 A0A182XFY5 A0A182JT48 A0A3B4Z3C1 A0A182YDV0 A0A0J7L6B5 A0A3P8WC14 A0A0A1WTE1 W8C5V8 A0A084W5X9 A0A182PLQ8 A0A182NTZ1 A0A182W6G3 A0A1Y1NKW7 A0A0K8VIM1 T1DQF3 A0A226EPG8 A0A182RDQ7 A0A182QRD4 A0A0P4WIT3 A0A182M7S6 A0A3B4TK66 A0A034WP28 A0A151WLL4 A0A0P5YRT6 A0A3B4XH03 A0A3P9BSK3 A0A3B1K2H6 E9IIQ0 A0A0P4WN66 A0A1Q3FFI2 A0A154P598 A0A3P8PQ78 A0A0P5V1G1 E9H6P3 I3IVW7 A0A060YB50
A0A1E1W1Y2 A0A2H1WVY8 S4PYG5 A0A0L7KY04 A0A336LMD6 A0A0K8TTA9 A0A023ELJ1 A0A182H1B5 U5EZL0 B4LQK4 A0A158P471 A0A195CTT7 A0A026WJJ8 B4GSH5 A0A1L8DTX1 Q29L16 F4WIQ6 A0A1J1I2E6 T1PDY6 A0A1L8EIV5 A0A151JRC9 A0A1L8EIP6 A0A1I8NNK4 A0A195FPA4 A0A0M3QTR3 B3MP76 A0A1B0GHY6 A0A3B0JNK3 B4JR23 A0A1B6D008 B4KFM6 B3N4D4 V5GVL0 A0A1W4UVL7 B4N0K6 B4I1A2 B4NW91 A0A2A3ECP4 A0A088AGR8 Q9VMW8 A0A1W4X3F3 B4Q361 A0A1S3SEA5 A0A060WHI1 A0A0L0CDH3 A0A182J0R7 A0A182IC72 A0A1S3N8K7 A0A0C9R4Z4 A0A182L0K0 A0A182UNM9 A0A182U4R2 Q7Q4F6 A0A2M4CV78 A0A1S4GY33 A0A3P8SND2 A0A2M4AK58 A0A3P8ZUS6 A0A182XFY5 A0A182JT48 A0A3B4Z3C1 A0A182YDV0 A0A0J7L6B5 A0A3P8WC14 A0A0A1WTE1 W8C5V8 A0A084W5X9 A0A182PLQ8 A0A182NTZ1 A0A182W6G3 A0A1Y1NKW7 A0A0K8VIM1 T1DQF3 A0A226EPG8 A0A182RDQ7 A0A182QRD4 A0A0P4WIT3 A0A182M7S6 A0A3B4TK66 A0A034WP28 A0A151WLL4 A0A0P5YRT6 A0A3B4XH03 A0A3P9BSK3 A0A3B1K2H6 E9IIQ0 A0A0P4WN66 A0A1Q3FFI2 A0A154P598 A0A3P8PQ78 A0A0P5V1G1 E9H6P3 I3IVW7 A0A060YB50
Ontologies
PANTHER
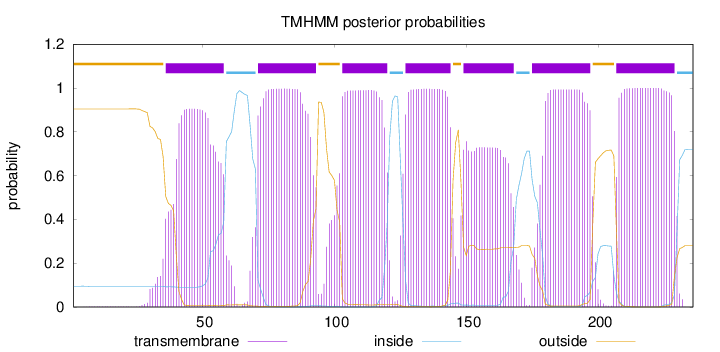
Topology
Subcellular location
Membrane
Length:
236
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
138.91521
Exp number, first 60 AAs:
18.52092
Total prob of N-in:
0.09529
POSSIBLE N-term signal
sequence
outside
1 - 35
TMhelix
36 - 58
inside
59 - 70
TMhelix
71 - 93
outside
94 - 102
TMhelix
103 - 120
inside
121 - 126
TMhelix
127 - 144
outside
145 - 148
TMhelix
149 - 168
inside
169 - 174
TMhelix
175 - 197
outside
198 - 206
TMhelix
207 - 229
inside
230 - 236
Population Genetic Test Statistics
Pi
176.661808
Theta
151.351788
Tajima's D
0.469788
CLR
0.036546
CSRT
0.501874906254687
Interpretation
Uncertain
