Gene
KWMTBOMO01787
Pre Gene Modal
BGIBMGA006153
Annotation
hypothetical_protein_OBRU01_16796?_partial_[Operophtera_brumata]
Full name
Tubulin glycylase 3A
Location in the cell
Nuclear Reliability : 2.617
Sequence
CDS
ATGTTAATACACGAAACAAAATTTGATATAAGGCAATATTATTTGATTACCAGTACATATCCTTTGATTATTTGGATATATAAAGACTGTTATTTGAAGTTCAGCTCTCAGAAATATAACCTTGGAAACTACCATGAATCTATACATTTAACTAACAACGCCGTACAAAGGAGATATATGAACTGTGAAGAGCGCCATCCTGATCTACCCAATAACAATATGTGGGATTTGGATACCTATAAAGATTATTTAAAGGCAATCGGCAAACCCTCGGTGTGGGATGATTTAATATATCCCGGTATGAAAAAGACAATCACAGGTGTGATGCTTAGTAGCCAAGAAACAATGCCGGCTTGCAAGAATCGCTTTGAACTGTACGGTTGCGATTTCCTACTGGACAAAGATTACAAGCCTTGGTTAATCGAAATCAATGCATGCCCTGATCTCAACAGTACCACACGAGTCACCGCGAAAATTTGTCCGGCCGTTTTAGCTGATGTCATTAAAGGTTTGATATAA
Protein
MLIHETKFDIRQYYLITSTYPLIIWIYKDCYLKFSSQKYNLGNYHESIHLTNNAVQRRYMNCEERHPDLPNNNMWDLDTYKDYLKAIGKPSVWDDLIYPGMKKTITGVMLSSQETMPACKNRFELYGCDFLLDKDYKPWLIEINACPDLNSTTRVTAKICPAVLADVIKGLI
Summary
Description
Polylycylase which modifies alpha- and beta-tubulin, generating side chains of glycine on the gamma-carboxyl groups of specific glutamate residues within the C-terminal tail of alpha- and beta-tubulin. Involved both in the side-chain initiation and elongation steps of the polyglycylation reaction by adding a single glycine chain to generate monoglycine side chains and by elongating monoglycine side chains to polyglycine side chains.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Cytoskeleton
Ligase
Microtubule
Nucleotide-binding
Reference proteome
Feature
chain Tubulin glycylase 3A
Uniprot
H9J9G0
A0A0L7KY04
A0A2H1WWR1
A0A212F7D8
A0A194R3R9
A0A194PMN9
+ More
A0A2A4K994 H9J9F9 A0A1Y1LE01 A0A1Y1LLK0 A0A2A4KA36 A0A194R2Y8 A0A212F7E8 A0A1B6L156 A0A1B0GHY1 A0A1B0GQ96 A0A0L0C2V8 A0A1B6IUI5 A0A1Y1LIV3 A0A1B6JCV0 A0A1B6LW88 A0A1I8MX16 A0A088AGQ7 A0A1I8PR05 A0A2A3EDF0 A0A1W4WKB8 A0A1B6J6Z4 A0A1J1IE64 A0A1A9ZFE1 A0A1A9VF55 A0A1B6MFR9 A0A1B6DSQ9 A0A1B0FAA3 A0A3L8DD51 B4LRK5 A0A1Y1LG93 A0A1A9W5H6 A0A1B6G8A6 A0A1B6FPT8 B4KL62 A0A0J9QY02 A0A0M9AAH7 A0A084VK39 Q9VM91 B4HXX2 B3N5W4 B4P018 B3MKL8 A0A1W4UBD6 A0A154P7K7 B4JB87 B0WKD5 A0A0M4E3Y7 A0A1B6H008 A0A1B6L0Z4 Q29P16 E1ZXT0 A0A139WHK4 B4GJX5 A0A0J7L6G8 A0A1B6IKE7 A0A1A9XQP7 A0A0K8T179 A0A026WJT5 E2B9Y4 A0A0K8T169 A0A3B0KIW8 Q7Q156 A0A1B0BN49 Q17NV1 A0A1S4EWD3 A0A182WRE8 A0A158P470 A0A1B6KRF3 B4N1E9 A0A232FAY4 A0A1I8IQ12 A0A267H1G7 A0A1I8HUU7 K7JAY6 A0A1W4W909 A0A267E3V8 F4WIQ5 A0A182LS70 W5JPW2 A0A182QWW9 A0A182U9C5 A0A182H0X3 A0A1I8GTN0 A0A182V6D4 A0A1B6FI59 A0A1B6EMD8 A0A267H6M3 A0A182JSX6 A0A182F296 A0A195BNU9 A0A182W8U6 A0A182HRC3 A0A182PIX4 A0A182RWT9
A0A2A4K994 H9J9F9 A0A1Y1LE01 A0A1Y1LLK0 A0A2A4KA36 A0A194R2Y8 A0A212F7E8 A0A1B6L156 A0A1B0GHY1 A0A1B0GQ96 A0A0L0C2V8 A0A1B6IUI5 A0A1Y1LIV3 A0A1B6JCV0 A0A1B6LW88 A0A1I8MX16 A0A088AGQ7 A0A1I8PR05 A0A2A3EDF0 A0A1W4WKB8 A0A1B6J6Z4 A0A1J1IE64 A0A1A9ZFE1 A0A1A9VF55 A0A1B6MFR9 A0A1B6DSQ9 A0A1B0FAA3 A0A3L8DD51 B4LRK5 A0A1Y1LG93 A0A1A9W5H6 A0A1B6G8A6 A0A1B6FPT8 B4KL62 A0A0J9QY02 A0A0M9AAH7 A0A084VK39 Q9VM91 B4HXX2 B3N5W4 B4P018 B3MKL8 A0A1W4UBD6 A0A154P7K7 B4JB87 B0WKD5 A0A0M4E3Y7 A0A1B6H008 A0A1B6L0Z4 Q29P16 E1ZXT0 A0A139WHK4 B4GJX5 A0A0J7L6G8 A0A1B6IKE7 A0A1A9XQP7 A0A0K8T179 A0A026WJT5 E2B9Y4 A0A0K8T169 A0A3B0KIW8 Q7Q156 A0A1B0BN49 Q17NV1 A0A1S4EWD3 A0A182WRE8 A0A158P470 A0A1B6KRF3 B4N1E9 A0A232FAY4 A0A1I8IQ12 A0A267H1G7 A0A1I8HUU7 K7JAY6 A0A1W4W909 A0A267E3V8 F4WIQ5 A0A182LS70 W5JPW2 A0A182QWW9 A0A182U9C5 A0A182H0X3 A0A1I8GTN0 A0A182V6D4 A0A1B6FI59 A0A1B6EMD8 A0A267H6M3 A0A182JSX6 A0A182F296 A0A195BNU9 A0A182W8U6 A0A182HRC3 A0A182PIX4 A0A182RWT9
EC Number
6.3.2.-
Pubmed
EMBL
BABH01004776
JTDY01004642
KOB67919.1
ODYU01011578
SOQ57406.1
AGBW02009882
+ More
OWR49655.1 KQ460855 KPJ11885.1 KQ459601 KPI93999.1 NWSH01000013 PCG80845.1 BABH01004775 GEZM01058134 JAV71879.1 GEZM01058136 JAV71877.1 PCG80846.1 KPJ11884.1 OWR49656.1 GEBQ01022535 JAT17442.1 AJWK01010432 AJVK01006597 JRES01000973 KNC26660.1 GECU01017186 JAS90520.1 GEZM01058137 JAV71875.1 GECU01010640 JAS97066.1 GEBQ01012035 JAT27942.1 KZ288283 PBC29514.1 GECU01012772 JAS94934.1 CVRI01000047 CRK98567.1 GEBQ01005238 JAT34739.1 GEDC01008582 JAS28716.1 CCAG010016184 QOIP01000010 RLU18082.1 CH940649 EDW63600.1 GEZM01058135 JAV71878.1 GECZ01011081 GECZ01005312 JAS58688.1 JAS64457.1 GECZ01017579 GECZ01004738 JAS52190.1 JAS65031.1 CH933807 EDW11723.1 CM002910 KMY88609.1 KQ435700 KOX80565.1 ATLV01014106 KE524943 KFB38333.1 AE014134 CH480818 EDW51902.1 CH954177 EDV59123.1 CM000157 EDW87845.1 CH902620 EDV31571.1 KQ434822 KZC07100.1 CH916368 EDW02892.1 DS231970 EDS29740.1 CP012523 ALC38434.1 GECZ01001763 JAS68006.1 GEBQ01022614 JAT17363.1 CH379058 EAL34477.2 GL435087 EFN74052.1 KQ971343 KYB27443.1 CH479184 EDW36941.1 LBMM01000547 KMQ98084.1 GECU01020318 JAS87388.1 GBRD01006538 JAG59283.1 KK107167 EZA56223.1 GL446618 EFN87487.1 GBRD01006539 JAG59282.1 OUUW01000010 SPP85706.1 AAAB01008980 EAA13905.3 JXJN01017195 CH477196 EAT48373.1 ADTU01008677 GEBQ01025960 JAT14017.1 CH963920 EDW78060.1 NNAY01000494 OXU28004.1 NIVC01000089 PAA91402.1 AAZX01007838 NIVC01002647 PAA56255.1 GL888177 EGI65851.1 AXCM01001818 ADMH02000503 ETN66181.1 AXCN02001993 JXUM01003334 JXUM01003335 JXUM01003336 JXUM01003337 JXUM01003338 JXUM01003339 JXUM01003340 JXUM01003341 KQ560156 KXJ84138.1 GECZ01019878 JAS49891.1 GECZ01030690 JAS39079.1 NIVC01000019 PAA93940.1 KQ976435 KYM87039.1 APCN01001680
OWR49655.1 KQ460855 KPJ11885.1 KQ459601 KPI93999.1 NWSH01000013 PCG80845.1 BABH01004775 GEZM01058134 JAV71879.1 GEZM01058136 JAV71877.1 PCG80846.1 KPJ11884.1 OWR49656.1 GEBQ01022535 JAT17442.1 AJWK01010432 AJVK01006597 JRES01000973 KNC26660.1 GECU01017186 JAS90520.1 GEZM01058137 JAV71875.1 GECU01010640 JAS97066.1 GEBQ01012035 JAT27942.1 KZ288283 PBC29514.1 GECU01012772 JAS94934.1 CVRI01000047 CRK98567.1 GEBQ01005238 JAT34739.1 GEDC01008582 JAS28716.1 CCAG010016184 QOIP01000010 RLU18082.1 CH940649 EDW63600.1 GEZM01058135 JAV71878.1 GECZ01011081 GECZ01005312 JAS58688.1 JAS64457.1 GECZ01017579 GECZ01004738 JAS52190.1 JAS65031.1 CH933807 EDW11723.1 CM002910 KMY88609.1 KQ435700 KOX80565.1 ATLV01014106 KE524943 KFB38333.1 AE014134 CH480818 EDW51902.1 CH954177 EDV59123.1 CM000157 EDW87845.1 CH902620 EDV31571.1 KQ434822 KZC07100.1 CH916368 EDW02892.1 DS231970 EDS29740.1 CP012523 ALC38434.1 GECZ01001763 JAS68006.1 GEBQ01022614 JAT17363.1 CH379058 EAL34477.2 GL435087 EFN74052.1 KQ971343 KYB27443.1 CH479184 EDW36941.1 LBMM01000547 KMQ98084.1 GECU01020318 JAS87388.1 GBRD01006538 JAG59283.1 KK107167 EZA56223.1 GL446618 EFN87487.1 GBRD01006539 JAG59282.1 OUUW01000010 SPP85706.1 AAAB01008980 EAA13905.3 JXJN01017195 CH477196 EAT48373.1 ADTU01008677 GEBQ01025960 JAT14017.1 CH963920 EDW78060.1 NNAY01000494 OXU28004.1 NIVC01000089 PAA91402.1 AAZX01007838 NIVC01002647 PAA56255.1 GL888177 EGI65851.1 AXCM01001818 ADMH02000503 ETN66181.1 AXCN02001993 JXUM01003334 JXUM01003335 JXUM01003336 JXUM01003337 JXUM01003338 JXUM01003339 JXUM01003340 JXUM01003341 KQ560156 KXJ84138.1 GECZ01019878 JAS49891.1 GECZ01030690 JAS39079.1 NIVC01000019 PAA93940.1 KQ976435 KYM87039.1 APCN01001680
Proteomes
UP000005204
UP000037510
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000092461 UP000092462 UP000037069 UP000095301 UP000005203 UP000095300 UP000242457 UP000192223 UP000183832 UP000092445 UP000078200 UP000092444 UP000279307 UP000008792 UP000091820 UP000009192 UP000053105 UP000030765 UP000000803 UP000001292 UP000008711 UP000002282 UP000007801 UP000192221 UP000076502 UP000001070 UP000002320 UP000092553 UP000001819 UP000000311 UP000007266 UP000008744 UP000036403 UP000092443 UP000053097 UP000008237 UP000268350 UP000007062 UP000092460 UP000008820 UP000076407 UP000005205 UP000007798 UP000215335 UP000095280 UP000215902 UP000002358 UP000007755 UP000075883 UP000000673 UP000075886 UP000075902 UP000069940 UP000249989 UP000075903 UP000075881 UP000069272 UP000078540 UP000075920 UP000075840 UP000075885 UP000075900
UP000092461 UP000092462 UP000037069 UP000095301 UP000005203 UP000095300 UP000242457 UP000192223 UP000183832 UP000092445 UP000078200 UP000092444 UP000279307 UP000008792 UP000091820 UP000009192 UP000053105 UP000030765 UP000000803 UP000001292 UP000008711 UP000002282 UP000007801 UP000192221 UP000076502 UP000001070 UP000002320 UP000092553 UP000001819 UP000000311 UP000007266 UP000008744 UP000036403 UP000092443 UP000053097 UP000008237 UP000268350 UP000007062 UP000092460 UP000008820 UP000076407 UP000005205 UP000007798 UP000215335 UP000095280 UP000215902 UP000002358 UP000007755 UP000075883 UP000000673 UP000075886 UP000075902 UP000069940 UP000249989 UP000075903 UP000075881 UP000069272 UP000078540 UP000075920 UP000075840 UP000075885 UP000075900
Interpro
SUPFAM
SSF47473
SSF47473
Gene 3D
CDD
ProteinModelPortal
H9J9G0
A0A0L7KY04
A0A2H1WWR1
A0A212F7D8
A0A194R3R9
A0A194PMN9
+ More
A0A2A4K994 H9J9F9 A0A1Y1LE01 A0A1Y1LLK0 A0A2A4KA36 A0A194R2Y8 A0A212F7E8 A0A1B6L156 A0A1B0GHY1 A0A1B0GQ96 A0A0L0C2V8 A0A1B6IUI5 A0A1Y1LIV3 A0A1B6JCV0 A0A1B6LW88 A0A1I8MX16 A0A088AGQ7 A0A1I8PR05 A0A2A3EDF0 A0A1W4WKB8 A0A1B6J6Z4 A0A1J1IE64 A0A1A9ZFE1 A0A1A9VF55 A0A1B6MFR9 A0A1B6DSQ9 A0A1B0FAA3 A0A3L8DD51 B4LRK5 A0A1Y1LG93 A0A1A9W5H6 A0A1B6G8A6 A0A1B6FPT8 B4KL62 A0A0J9QY02 A0A0M9AAH7 A0A084VK39 Q9VM91 B4HXX2 B3N5W4 B4P018 B3MKL8 A0A1W4UBD6 A0A154P7K7 B4JB87 B0WKD5 A0A0M4E3Y7 A0A1B6H008 A0A1B6L0Z4 Q29P16 E1ZXT0 A0A139WHK4 B4GJX5 A0A0J7L6G8 A0A1B6IKE7 A0A1A9XQP7 A0A0K8T179 A0A026WJT5 E2B9Y4 A0A0K8T169 A0A3B0KIW8 Q7Q156 A0A1B0BN49 Q17NV1 A0A1S4EWD3 A0A182WRE8 A0A158P470 A0A1B6KRF3 B4N1E9 A0A232FAY4 A0A1I8IQ12 A0A267H1G7 A0A1I8HUU7 K7JAY6 A0A1W4W909 A0A267E3V8 F4WIQ5 A0A182LS70 W5JPW2 A0A182QWW9 A0A182U9C5 A0A182H0X3 A0A1I8GTN0 A0A182V6D4 A0A1B6FI59 A0A1B6EMD8 A0A267H6M3 A0A182JSX6 A0A182F296 A0A195BNU9 A0A182W8U6 A0A182HRC3 A0A182PIX4 A0A182RWT9
A0A2A4K994 H9J9F9 A0A1Y1LE01 A0A1Y1LLK0 A0A2A4KA36 A0A194R2Y8 A0A212F7E8 A0A1B6L156 A0A1B0GHY1 A0A1B0GQ96 A0A0L0C2V8 A0A1B6IUI5 A0A1Y1LIV3 A0A1B6JCV0 A0A1B6LW88 A0A1I8MX16 A0A088AGQ7 A0A1I8PR05 A0A2A3EDF0 A0A1W4WKB8 A0A1B6J6Z4 A0A1J1IE64 A0A1A9ZFE1 A0A1A9VF55 A0A1B6MFR9 A0A1B6DSQ9 A0A1B0FAA3 A0A3L8DD51 B4LRK5 A0A1Y1LG93 A0A1A9W5H6 A0A1B6G8A6 A0A1B6FPT8 B4KL62 A0A0J9QY02 A0A0M9AAH7 A0A084VK39 Q9VM91 B4HXX2 B3N5W4 B4P018 B3MKL8 A0A1W4UBD6 A0A154P7K7 B4JB87 B0WKD5 A0A0M4E3Y7 A0A1B6H008 A0A1B6L0Z4 Q29P16 E1ZXT0 A0A139WHK4 B4GJX5 A0A0J7L6G8 A0A1B6IKE7 A0A1A9XQP7 A0A0K8T179 A0A026WJT5 E2B9Y4 A0A0K8T169 A0A3B0KIW8 Q7Q156 A0A1B0BN49 Q17NV1 A0A1S4EWD3 A0A182WRE8 A0A158P470 A0A1B6KRF3 B4N1E9 A0A232FAY4 A0A1I8IQ12 A0A267H1G7 A0A1I8HUU7 K7JAY6 A0A1W4W909 A0A267E3V8 F4WIQ5 A0A182LS70 W5JPW2 A0A182QWW9 A0A182U9C5 A0A182H0X3 A0A1I8GTN0 A0A182V6D4 A0A1B6FI59 A0A1B6EMD8 A0A267H6M3 A0A182JSX6 A0A182F296 A0A195BNU9 A0A182W8U6 A0A182HRC3 A0A182PIX4 A0A182RWT9
PDB
5VLQ
E-value=1.742e-44,
Score=447
Ontologies
KEGG
GO
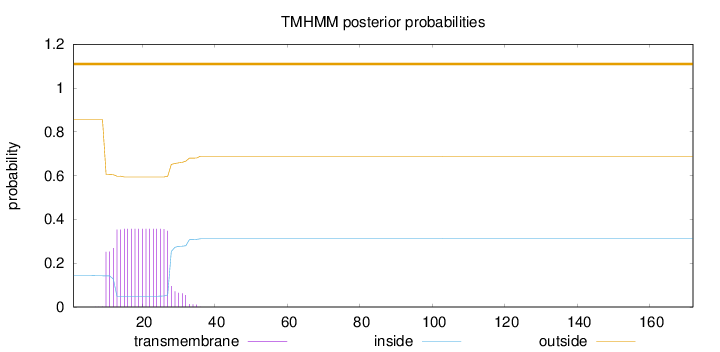
Topology
Subcellular location
Length:
172
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.49663999999999
Exp number, first 60 AAs:
6.49584
Total prob of N-in:
0.14355
outside
1 - 172
Population Genetic Test Statistics
Pi
310.206653
Theta
212.03484
Tajima's D
1.45769
CLR
0
CSRT
0.781110944452777
Interpretation
Uncertain
