Gene
KWMTBOMO01784
Pre Gene Modal
BGIBMGA006151
Annotation
integrin_beta4_[Bombyx_mori]
Full name
Integrin beta
+ More
Integrin beta-3
Integrin beta-6
Integrin beta-3
Integrin beta-6
Alternative Name
Platelet membrane glycoprotein IIIa
Location in the cell
PlasmaMembrane Reliability : 4.938
Sequence
CDS
ATGATTACTCAACACTGCTGGCCTGAATTCTCGGATGCCTGTGCGAAAAACTTCGAGGAACTACCGTCCGCGATGTTAAGAAGCAATAACGGGACGAAGTTACTGAGCATTAACTACATTGAACAATCCGTGCGGCCGTCTTCGCAATATGAACTGACCATAGAATACAACGCGGCGACTCAATCTCAGATAAAAATAGAAATCGTAAACTCGACGCAGACTAGGAATCTTAAATTTTCTGACACAACGAAATGCGAAAATTCTGAATGCACTGCAAAGATTGCGGTCAGGCCATTGGAAACGTTCTGCACGTCTTCCGGTAGTCCTTACGAATACGTATACGCGAACATTTTCTTTGTGGACCTGAACGAATCGACATTAGTGAAGTATTTGATTTCGTGCGCGTGTCCTTGTTCCGCCCAACCCGAACCAAACTCGGAGACGTGCCACAAGAACGGACAATTAATTTGTGGTATTTGCGAATGCTACGACGGATGGTCTGGAAGTTTCTGCAACGAAGCAGCTCAGACTCCAATAACGCCTGTTGTACCTACAATTGTTGAGAAGAAATCACCGAAATGTGAATATCACGGCCATTGGCTGTCAAGCGGAATTTGCGATTGCTTTAAAGGATGGACGGGAAAGAACTGCAACGTACTGAACTGCGGACGCAGACGTGGAGATATCGGCTGCAGTCGAACCAGTGAACTTGGGGAATGCTCAGGGAACGGTGAGTGCGACGCTTGCGGCGACGCCTGCGTCTGCAAAACCGACATCGAAGGGGCCCAGTACTTAGAAGCGGACAACAACTGCGTGGACCTGTGCATGACTGTTGAACAGTGCAAGGATTGTTTATCTAGCAACGGGCCGATGCAGTGTTCGAACTGCAACATTGTGAAGGACGCGTACAACCGTACCGTCTCGGAAGAGCTAGACTATTTAAACAGGAGAGTGTGGATGCATTGCAGCTACATCAACGTCACGACTGAGTGCAAAGTCTTGTATTTGGCGATGAGAGAAAACGAAGTTGTGCATATAATGATTGAGGACTATTGCGAGGTGTCAACGCCGGCCGGTCTGTTCGCGTCGCCGGTGACGTCGGTACTATTTGGACTACTAGCCGTGATATGCGCGGCGTCGCTAGTCGCTGTGGCCGTGTGGAAGTACTTCAATCGCATCCCAGCACAGGACTTGATAAAGCCAGGCTACATGAGCGTGGAGGCGGCCACAAGCACCGTACAGAACCCGGCCTACGTGACGCCGATCTCTTCGTTCATCAACCCTACGTACCAAAAATGCTGA
Protein
MITQHCWPEFSDACAKNFEELPSAMLRSNNGTKLLSINYIEQSVRPSSQYELTIEYNAATQSQIKIEIVNSTQTRNLKFSDTTKCENSECTAKIAVRPLETFCTSSGSPYEYVYANIFFVDLNESTLVKYLISCACPCSAQPEPNSETCHKNGQLICGICECYDGWSGSFCNEAAQTPITPVVPTIVEKKSPKCEYHGHWLSSGICDCFKGWTGKNCNVLNCGRRRGDIGCSRTSELGECSGNGECDACGDACVCKTDIEGAQYLEADNNCVDLCMTVEQCKDCLSSNGPMQCSNCNIVKDAYNRTVSEELDYLNRRVWMHCSYINVTTECKVLYLAMRENEVVHIMIEDYCEVSTPAGLFASPVTSVLFGLLAVICAASLVAVAVWKYFNRIPAQDLIKPGYMSVEAATSTVQNPAYVTPISSFINPTYQKC
Summary
Description
Integrin alpha-V/beta-3 (ITGAV:ITGB3) is a receptor for cytotactin, fibronectin, laminin, matrix metalloproteinase-2, osteopontin, osteomodulin, prothrombin, thrombospondin, vitronectin and von Willebrand factor. Integrin alpha-IIB/beta-3 (ITGA2B:ITGB3) is a receptor for fibronectin, fibrinogen, plasminogen, prothrombin, thrombospondin and vitronectin. Integrins alpha-IIB/beta-3 and alpha-V/beta-3 recognize the sequence R-G-D in a wide array of ligands. Integrin alpha-IIB/beta-3 recognizes the sequence H-H-L-G-G-G-A-K-Q-A-G-D-V in fibrinogen gamma chain. Following activation integrin alpha-IIB/beta-3 brings about platelet/platelet interaction through binding of soluble fibrinogen. This step leads to rapid platelet aggregation which physically plugs ruptured endothelial surfaces. Fibrinogen binding enhances SELP expression in activated platelets (PubMed:19332769). ITGAV:ITGB3 binds to fractalkine (CX3CL1) and acts as its coreceptor in CX3CR1-dependent fractalkine signaling. ITGAV:ITGB3 binds to NRG1 (via EGF domain) and this binding is essential for NRG1-ERBB signaling. ITGAV:ITGB3 binds to FGF1 and this binding is essential for FGF1 signaling. ITGAV:ITGB3 binds to FGF2 and this binding is essential for FGF2 signaling (By similarity). ITGAV:ITGB3 binds to IGF1 and this binding is essential for IGF1 signaling (By similarity). ITGAV:ITGB3 binds to IGF2 and this binding is essential for IGF2 signaling (By similarity). ITGAV:ITGB3 binds to IL1B and this binding is essential for IL1B signaling (By similarity). ITGAV:ITGB3 binds to PLA2G2A via a site (site 2) which is distinct from the classical ligand-binding site (site 1) and this induces integrin conformational changes and enhanced ligand binding to site 1 (By similarity). ITGAV:ITGB3 acts as a receptor for fibrillin-1 (FBN1) and mediates R-G-D-dependent cell adhesion to FBN1 (By similarity).
Integrin alpha-V:beta-6 (ITGAV:ITGB6) is a receptor for fibronectin and cytotactin (PubMed:17545607, PubMed:17158881). It recognizes the sequence R-G-D in its ligands (PubMed:17545607, PubMed:17158881). Internalisation of integrin alpha-V/beta-6 via clathrin-mediated endocytosis promotes carcinoma cell invasion (PubMed:17545607, PubMed:17158881). ITGAV:ITGB6 acts as a receptor for fibrillin-1 (FBN1) and mediates R-G-D-dependent cell adhesion to FBN1 (PubMed:17158881). Integrin alpha-V:beta-6 (ITGAV:ITGB6) mediates R-G-D-dependent release of transforming growth factor beta-1 (TGF-beta-1) from regulatory Latency-associated peptide (LAP), thereby playing a key role in TGF-beta-1 activation (PubMed:15184403, PubMed:22278742, PubMed:28117447).
(Microbial infection) Integrin ITGAV:ITGB6 acts as a receptor for Coxsackievirus A9 and Coxsackievirus B1.
(Microbial infection) Integrin ITGAV:ITGB6 acts as a receptor for Herpes simplex virus-1/HHV-1 (PubMed:24367260).
Integrin alpha-V:beta-6 (ITGAV:ITGB6) is a receptor for fibronectin and cytotactin (PubMed:17545607, PubMed:17158881). It recognizes the sequence R-G-D in its ligands (PubMed:17545607, PubMed:17158881). Internalisation of integrin alpha-V/beta-6 via clathrin-mediated endocytosis promotes carcinoma cell invasion (PubMed:17545607, PubMed:17158881). ITGAV:ITGB6 acts as a receptor for fibrillin-1 (FBN1) and mediates R-G-D-dependent cell adhesion to FBN1 (PubMed:17158881). Integrin alpha-V:beta-6 (ITGAV:ITGB6) mediates R-G-D-dependent release of transforming growth factor beta-1 (TGF-beta-1) from regulatory Latency-associated peptide (LAP), thereby playing a key role in TGF-beta-1 activation (PubMed:15184403, PubMed:22278742, PubMed:28117447).
(Microbial infection) Integrin ITGAV:ITGB6 acts as a receptor for Coxsackievirus A9 and Coxsackievirus B1.
(Microbial infection) Integrin ITGAV:ITGB6 acts as a receptor for Herpes simplex virus-1/HHV-1 (PubMed:24367260).
Subunit
Heterodimer of an alpha and a beta subunit (By similarity). Beta-3 (ITGB3) associates with either alpha-IIB (ITGA2B) or alpha-V (ITGAV). Interacts with FLNB and COMP (By similarity). Interacts with PDIA6 following platelet stimulation (By similarity). Interacts with SYK; upon activation by ITGB3 promotes platelet adhesion (By similarity). Interacts with MYO10 (By similarity). Interacts with DAB2 (PubMed:12606711). Interacts with FERMT2 (PubMed:18483218). Integrin ITGAV:ITGB3 interacts with FBLN5 (via N-terminus) (PubMed:11805835). Interacts with EMP2; regulates the levels of the heterodimer ITGA5:ITGB3 integrin expression on the plasma membrane (By similarity). ITGAV:ITGB3 interacts with CCN3 (By similarity). ITGAV:ITGB3 interacts with AGRA2 (By similarity). ITGAV:ITGB3 is found in a ternary complex with CX3CR1 and CX3CL1. ITGAV:ITGB3 is found in a ternary complex with NRG1 and ERBB3. ITGAV:ITGB3 is found in a ternary complex with FGF1 and FGFR1. ITGAV:ITGB3 interacts with FGF2; it is likely that FGF2 can simultaneously bind ITGAV:ITGB3 and FGF receptors (By similarity). ITGAV:ITGB3 binds to IL1B (By similarity). ITGAV:ITGB3 is found in a ternary complex with IGF1 and IGF1R (By similarity). ITGAV:ITGB3 interacts with IGF2 (By similarity). ITGAV:ITGB3 interacts with FBN1 (By similarity). ITGAV:ITGB3 interacts with CD9, CD81 and CD151 (via second extracellular domain) (By similarity). Interacts (via the allosteric site (site 2)) with CXCL12 in a CXCR4-independent manner (By similarity). Interacts with MXRA8/DICAM; the interaction inhibits ITGAV:ITGB3 heterodimer formation (PubMed:22492581). ITGAV:ITGB3 interacts with PTN. Forms a complex with PTPRZ1 and PTN that stimulates endothelial cell migration through ITGB3 Tyr-772 phosphorylation (By similarity).
Heterodimer of an alpha and a beta subunit (PubMed:11807098, PubMed:17545607, PubMed:17158881). Interacts with FLNB (PubMed:11807098). Interacts with HAX1 (PubMed:17545607). ITGAV:ITGB6 interacts with FBN1 (PubMed:17158881). ITGAV:ITGB6 interacts with TGFB1 (PubMed:22278742, PubMed:28117447).
(Microbial infection) Integrin ITGAV:ITGB6 interacts with coxsackievirus A9, coxsackievirus B1 capsid proteins (PubMed:9426447, PubMed:15194773).
(Microbial infection) Integrin ITGAV:ITGB6 interacts with herpes simplex virus-1/HHV-1 gH:gL proteins.
Heterodimer of an alpha and a beta subunit (PubMed:11807098, PubMed:17545607, PubMed:17158881). Interacts with FLNB (PubMed:11807098). Interacts with HAX1 (PubMed:17545607). ITGAV:ITGB6 interacts with FBN1 (PubMed:17158881). ITGAV:ITGB6 interacts with TGFB1 (PubMed:22278742, PubMed:28117447).
(Microbial infection) Integrin ITGAV:ITGB6 interacts with coxsackievirus A9, coxsackievirus B1 capsid proteins (PubMed:9426447, PubMed:15194773).
(Microbial infection) Integrin ITGAV:ITGB6 interacts with herpes simplex virus-1/HHV-1 gH:gL proteins.
Similarity
Belongs to the integrin beta chain family.
Keywords
3D-structure
Cell adhesion
Cell junction
Cell membrane
Cell projection
Complete proteome
Disulfide bond
Glycoprotein
Integrin
Membrane
Phosphoprotein
Receptor
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Alternative splicing
Amelogenesis imperfecta
Disease mutation
Host cell receptor for virus entry
Host-virus interaction
Polymorphism
Feature
chain Integrin beta
splice variant In isoform 2.
sequence variant In AI1H; dbSNP:rs140015315.
splice variant In isoform 2.
sequence variant In AI1H; dbSNP:rs140015315.
Uniprot
A0A076JU43
H9J9F8
A0A1S6PU27
A0A2H1V7R3
A0A2A4KAT3
A0A212F7G3
+ More
A0A212FB72 A0A194R7N4 A0A194PSA2 D6X0D6 A0A336KRK3 F6SRR1 A0A336MVT2 A0A1S3GJ18 A0A2K5LTD5 A0A2K5J5N2 A0A2K6KCB9 A0A2K5LTE2 A0A2K6BAY6 A0A2K5J5H3 A0A2K6KC64 A0A2K6Q2S9 A0A096P692 A0A2K5LTC9 A0A2K6BAZ5 A0A2K5YHH7 A0A2I3MIT0 A0A2K5LTD1 G7NJ66 G7PV32 A0A2K5J5M1 A0A0D9QUZ5 F7FG54 A0A2K5WY82 A0A2I3NH66 B7ZQZ8 Q6GP06 Q07012 A0A2Y9ED82 H0XE40 A0A340X9R2 A0A340X5H4 A0A2Y9H9C9 A0A2Y9H988 G1M853 A0A3P8YQD8 B5APS2 A0A3P8YNF4 A0A2U3VC82 A0A1L8DL40 A0A383ZL92 A0A1L8DMT3 A0A1L8DMF7 G1LCS5 A0A1L8DMR9 D2H505 W8AY85 A0A1L8DMS5 A0A1L8DMI8 A0A1L8DMJ4 A0A0B5H7U8 S4RSM3 A0A087WXP3 O54890 B4E2B8 G1TSS7 F7FW35 J9P3C1 P18564 A0A3B3XFV2 A0A2I3TGA0 K7DU23 Q9TUN3 K7DN27 A0A2M4ACS8 A0A3Q0CI27 E9PEE8 A0A2M4ACZ9 H2QDA1 A0A2Y9KBB9 A8K2N5 G1T7R4 A0A2I3RVJ2 A0A3P9Q7K9 G3HGD1
A0A212FB72 A0A194R7N4 A0A194PSA2 D6X0D6 A0A336KRK3 F6SRR1 A0A336MVT2 A0A1S3GJ18 A0A2K5LTD5 A0A2K5J5N2 A0A2K6KCB9 A0A2K5LTE2 A0A2K6BAY6 A0A2K5J5H3 A0A2K6KC64 A0A2K6Q2S9 A0A096P692 A0A2K5LTC9 A0A2K6BAZ5 A0A2K5YHH7 A0A2I3MIT0 A0A2K5LTD1 G7NJ66 G7PV32 A0A2K5J5M1 A0A0D9QUZ5 F7FG54 A0A2K5WY82 A0A2I3NH66 B7ZQZ8 Q6GP06 Q07012 A0A2Y9ED82 H0XE40 A0A340X9R2 A0A340X5H4 A0A2Y9H9C9 A0A2Y9H988 G1M853 A0A3P8YQD8 B5APS2 A0A3P8YNF4 A0A2U3VC82 A0A1L8DL40 A0A383ZL92 A0A1L8DMT3 A0A1L8DMF7 G1LCS5 A0A1L8DMR9 D2H505 W8AY85 A0A1L8DMS5 A0A1L8DMI8 A0A1L8DMJ4 A0A0B5H7U8 S4RSM3 A0A087WXP3 O54890 B4E2B8 G1TSS7 F7FW35 J9P3C1 P18564 A0A3B3XFV2 A0A2I3TGA0 K7DU23 Q9TUN3 K7DN27 A0A2M4ACS8 A0A3Q0CI27 E9PEE8 A0A2M4ACZ9 H2QDA1 A0A2Y9KBB9 A8K2N5 G1T7R4 A0A2I3RVJ2 A0A3P9Q7K9 G3HGD1
Pubmed
25064490
19121390
28232041
22118469
26354079
18362917
+ More
19820115 20431018 25362486 22002653 17431167 27762356 7693527 20010809 25069045 19443046 24495485 15815621 16141072 19468303 15489334 11805835 12606711 18483218 19332769 21183079 22492581 21993624 17495919 16341006 2365683 14702039 1382574 9426447 11807098 15184403 15194773 17545607 17158881 22278742 24367260 24319098 24305999 28117447 16136131 26319212 21804562
19820115 20431018 25362486 22002653 17431167 27762356 7693527 20010809 25069045 19443046 24495485 15815621 16141072 19468303 15489334 11805835 12606711 18483218 19332769 21183079 22492581 21993624 17495919 16341006 2365683 14702039 1382574 9426447 11807098 15184403 15194773 17545607 17158881 22278742 24367260 24319098 24305999 28117447 16136131 26319212 21804562
EMBL
KJ511857
AII79416.1
BABH01004757
BABH01004758
BABH01004759
BABH01004760
+ More
KY346540 AQV03248.1 ODYU01001118 SOQ36885.1 NWSH01000013 PCG80850.1 AGBW02009882 OWR49658.1 AGBW02009374 OWR50968.1 KQ460855 KPJ11881.1 KQ459601 KPI94005.1 KQ971372 EFA10689.2 UFQS01000673 UFQT01000673 SSX06025.1 SSX26382.1 AAMC01124678 AAMC01124679 UFQS01003187 UFQT01003187 SSX15340.1 SSX34714.1 AHZZ02010501 CM001268 EHH25088.1 AQIA01028217 CM001291 EHH58231.1 AQIB01117870 JSUE03016688 BC169987 BC169991 CM004482 AAI69987.1 OCT62291.1 BC073343 AAH73343.1 L13591 AAA17427.1 AAQR03042280 AAQR03042281 ACTA01040108 EU867790 ACF93732.1 GFDF01006905 JAV07179.1 GFDF01006410 JAV07674.1 GFDF01006529 JAV07555.1 ACTA01050688 ACTA01058688 ACTA01066688 ACTA01074688 GFDF01006409 JAV07675.1 GL192494 EFB14276.1 GAMC01020742 GAMC01020741 GAMC01020740 GAMC01020739 GAMC01020738 JAB85816.1 GFDF01006407 JAV07677.1 GFDF01006408 JAV07676.1 GFDF01006406 JAV07678.1 KJ535130 AJF38203.1 AC080166 AC092153 AF026509 AK157958 AL603709 BX000996 CH466558 BC125518 BC125520 AK304203 BAG65080.1 AAGW02039896 AAEX03017055 M35198 AK313944 CH471058 BC121178 S49380 AACZ04071172 AC185328 GABE01000473 GABE01000472 GABE01000471 GABE01000469 JAA44267.1 AF170529 AAD51955.1 GABE01000470 JAA44269.1 GGFK01005275 MBW38596.1 GGFK01005281 MBW38602.1 AK290300 BAF82989.1 AAGW02013727 AAGW02013728 JH000347 EGV96758.1
KY346540 AQV03248.1 ODYU01001118 SOQ36885.1 NWSH01000013 PCG80850.1 AGBW02009882 OWR49658.1 AGBW02009374 OWR50968.1 KQ460855 KPJ11881.1 KQ459601 KPI94005.1 KQ971372 EFA10689.2 UFQS01000673 UFQT01000673 SSX06025.1 SSX26382.1 AAMC01124678 AAMC01124679 UFQS01003187 UFQT01003187 SSX15340.1 SSX34714.1 AHZZ02010501 CM001268 EHH25088.1 AQIA01028217 CM001291 EHH58231.1 AQIB01117870 JSUE03016688 BC169987 BC169991 CM004482 AAI69987.1 OCT62291.1 BC073343 AAH73343.1 L13591 AAA17427.1 AAQR03042280 AAQR03042281 ACTA01040108 EU867790 ACF93732.1 GFDF01006905 JAV07179.1 GFDF01006410 JAV07674.1 GFDF01006529 JAV07555.1 ACTA01050688 ACTA01058688 ACTA01066688 ACTA01074688 GFDF01006409 JAV07675.1 GL192494 EFB14276.1 GAMC01020742 GAMC01020741 GAMC01020740 GAMC01020739 GAMC01020738 JAB85816.1 GFDF01006407 JAV07677.1 GFDF01006408 JAV07676.1 GFDF01006406 JAV07678.1 KJ535130 AJF38203.1 AC080166 AC092153 AF026509 AK157958 AL603709 BX000996 CH466558 BC125518 BC125520 AK304203 BAG65080.1 AAGW02039896 AAEX03017055 M35198 AK313944 CH471058 BC121178 S49380 AACZ04071172 AC185328 GABE01000473 GABE01000472 GABE01000471 GABE01000469 JAA44267.1 AF170529 AAD51955.1 GABE01000470 JAA44269.1 GGFK01005275 MBW38596.1 GGFK01005281 MBW38602.1 AK290300 BAF82989.1 AAGW02013727 AAGW02013728 JH000347 EGV96758.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
+ More
UP000008143 UP000081671 UP000233060 UP000233080 UP000233180 UP000233120 UP000233200 UP000028761 UP000233140 UP000009130 UP000233100 UP000029965 UP000006718 UP000186698 UP000248484 UP000005225 UP000265300 UP000248481 UP000008912 UP000265140 UP000245340 UP000261681 UP000245300 UP000005640 UP000000589 UP000001811 UP000002280 UP000002254 UP000261480 UP000002277 UP000189706 UP000248482 UP000242638 UP000001075
UP000008143 UP000081671 UP000233060 UP000233080 UP000233180 UP000233120 UP000233200 UP000028761 UP000233140 UP000009130 UP000233100 UP000029965 UP000006718 UP000186698 UP000248484 UP000005225 UP000265300 UP000248481 UP000008912 UP000265140 UP000245340 UP000261681 UP000245300 UP000005640 UP000000589 UP000001811 UP000002280 UP000002254 UP000261480 UP000002277 UP000189706 UP000248482 UP000242638 UP000001075
Pfam
Interpro
IPR040622
I-EGF_1
+ More
IPR015812 Integrin_bsu
IPR014836 Integrin_bsu_cyt_dom
IPR036465 vWFA_dom_sf
IPR032695 Integrin_dom_sf
IPR013111 EGF_extracell
IPR033760 Integrin_beta_N
IPR002369 Integrin_bsu_VWA
IPR012896 Integrin_bsu_tail
IPR036349 Integrin_bsu_tail_dom_sf
IPR027068 Integrin_beta-3
IPR016201 PSI
IPR027067 Integrin_beta-5
IPR002035 VWF_A
IPR015436 Integrin_bsu-6
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR015812 Integrin_bsu
IPR014836 Integrin_bsu_cyt_dom
IPR036465 vWFA_dom_sf
IPR032695 Integrin_dom_sf
IPR013111 EGF_extracell
IPR033760 Integrin_beta_N
IPR002369 Integrin_bsu_VWA
IPR012896 Integrin_bsu_tail
IPR036349 Integrin_bsu_tail_dom_sf
IPR027068 Integrin_beta-3
IPR016201 PSI
IPR027067 Integrin_beta-5
IPR002035 VWF_A
IPR015436 Integrin_bsu-6
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
Gene 3D
CDD
ProteinModelPortal
A0A076JU43
H9J9F8
A0A1S6PU27
A0A2H1V7R3
A0A2A4KAT3
A0A212F7G3
+ More
A0A212FB72 A0A194R7N4 A0A194PSA2 D6X0D6 A0A336KRK3 F6SRR1 A0A336MVT2 A0A1S3GJ18 A0A2K5LTD5 A0A2K5J5N2 A0A2K6KCB9 A0A2K5LTE2 A0A2K6BAY6 A0A2K5J5H3 A0A2K6KC64 A0A2K6Q2S9 A0A096P692 A0A2K5LTC9 A0A2K6BAZ5 A0A2K5YHH7 A0A2I3MIT0 A0A2K5LTD1 G7NJ66 G7PV32 A0A2K5J5M1 A0A0D9QUZ5 F7FG54 A0A2K5WY82 A0A2I3NH66 B7ZQZ8 Q6GP06 Q07012 A0A2Y9ED82 H0XE40 A0A340X9R2 A0A340X5H4 A0A2Y9H9C9 A0A2Y9H988 G1M853 A0A3P8YQD8 B5APS2 A0A3P8YNF4 A0A2U3VC82 A0A1L8DL40 A0A383ZL92 A0A1L8DMT3 A0A1L8DMF7 G1LCS5 A0A1L8DMR9 D2H505 W8AY85 A0A1L8DMS5 A0A1L8DMI8 A0A1L8DMJ4 A0A0B5H7U8 S4RSM3 A0A087WXP3 O54890 B4E2B8 G1TSS7 F7FW35 J9P3C1 P18564 A0A3B3XFV2 A0A2I3TGA0 K7DU23 Q9TUN3 K7DN27 A0A2M4ACS8 A0A3Q0CI27 E9PEE8 A0A2M4ACZ9 H2QDA1 A0A2Y9KBB9 A8K2N5 G1T7R4 A0A2I3RVJ2 A0A3P9Q7K9 G3HGD1
A0A212FB72 A0A194R7N4 A0A194PSA2 D6X0D6 A0A336KRK3 F6SRR1 A0A336MVT2 A0A1S3GJ18 A0A2K5LTD5 A0A2K5J5N2 A0A2K6KCB9 A0A2K5LTE2 A0A2K6BAY6 A0A2K5J5H3 A0A2K6KC64 A0A2K6Q2S9 A0A096P692 A0A2K5LTC9 A0A2K6BAZ5 A0A2K5YHH7 A0A2I3MIT0 A0A2K5LTD1 G7NJ66 G7PV32 A0A2K5J5M1 A0A0D9QUZ5 F7FG54 A0A2K5WY82 A0A2I3NH66 B7ZQZ8 Q6GP06 Q07012 A0A2Y9ED82 H0XE40 A0A340X9R2 A0A340X5H4 A0A2Y9H9C9 A0A2Y9H988 G1M853 A0A3P8YQD8 B5APS2 A0A3P8YNF4 A0A2U3VC82 A0A1L8DL40 A0A383ZL92 A0A1L8DMT3 A0A1L8DMF7 G1LCS5 A0A1L8DMR9 D2H505 W8AY85 A0A1L8DMS5 A0A1L8DMI8 A0A1L8DMJ4 A0A0B5H7U8 S4RSM3 A0A087WXP3 O54890 B4E2B8 G1TSS7 F7FW35 J9P3C1 P18564 A0A3B3XFV2 A0A2I3TGA0 K7DU23 Q9TUN3 K7DN27 A0A2M4ACS8 A0A3Q0CI27 E9PEE8 A0A2M4ACZ9 H2QDA1 A0A2Y9KBB9 A8K2N5 G1T7R4 A0A2I3RVJ2 A0A3P9Q7K9 G3HGD1
PDB
6BXJ
E-value=1.55108e-11,
Score=168
Ontologies
GO
GO:0016021
GO:0007229
GO:0009986
GO:0033627
GO:0007160
GO:0005925
GO:0005178
GO:0008305
GO:0016477
GO:0007155
GO:0001968
GO:0070527
GO:0035866
GO:0001954
GO:0010763
GO:0007044
GO:0043277
GO:0048008
GO:0036120
GO:2000406
GO:1900026
GO:0098978
GO:0099699
GO:1903053
GO:0061097
GO:0032956
GO:0005080
GO:0048146
GO:0010628
GO:0009897
GO:0071133
GO:0033690
GO:0016324
GO:0038023
GO:0034684
GO:0043149
GO:0035987
GO:0090136
GO:0007179
GO:0005654
GO:0030054
GO:0005813
GO:0032587
GO:0042470
GO:0099149
GO:0031528
GO:0035868
GO:0042802
GO:0032147
GO:0050839
GO:0010888
GO:0050731
GO:0050840
GO:0048858
GO:0034446
GO:0005886
GO:0048661
GO:0050748
GO:0045766
GO:0030168
GO:0010745
GO:0034679
GO:0070374
GO:0072126
GO:0007566
GO:0010595
GO:0038027
GO:0014911
GO:0031527
GO:0070051
GO:0032369
GO:0034683
GO:0043235
GO:0016020
GO:0033630
GO:0045672
GO:1990314
GO:0035867
GO:0003756
GO:0001938
GO:0031589
GO:0045715
GO:0014823
GO:0046718
GO:0014909
GO:0071260
GO:0030335
GO:0019899
GO:0009314
GO:1900731
GO:0034113
GO:0008277
GO:0051279
GO:0030334
GO:0043184
GO:0002020
GO:0005634
GO:0002687
GO:0060548
GO:0045780
GO:0050919
GO:0031258
GO:0035690
GO:0034685
GO:0006954
GO:0038044
GO:1901388
GO:0001618
GO:0030198
GO:0005509
Topology
Subcellular location
Membrane
Cell membrane
Cell projection
Lamellipodium membrane
Cell junction
Focal adhesion
Cell membrane
Cell projection
Lamellipodium membrane
Cell junction
Focal adhesion
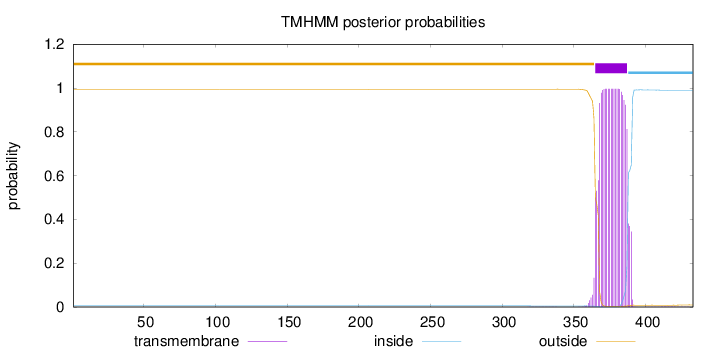
Length:
433
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.55924
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00534
outside
1 - 364
TMhelix
365 - 387
inside
388 - 433
Population Genetic Test Statistics
Pi
237.163594
Theta
195.07975
Tajima's D
0.498285
CLR
0.471868
CSRT
0.508324583770811
Interpretation
Uncertain
