Gene
KWMTBOMO01780
Annotation
PREDICTED:_integrin_beta-PS-like_[Bombyx_mori]
Full name
Integrin beta
Location in the cell
Extracellular Reliability : 1.427 Nuclear Reliability : 1.617
Sequence
CDS
ATGAATAGCAGTGATTCGCTTGAAGACCCTTTGGAACCGAGTTGGAATTTATGCCCAGAACTGAGGATGGAAAACGGCTGCTATACGACTTTTGCATATAAATATCAAGATGGTTACGGGATCGAATTATTGATACAGAAAAACAAGAATTGCGATGAGAATATTTTAAAGCTGGGCGTGATAGTATCTGTAACCGTCATCTTGGTAGGGATCGGAACATTGGTTGTTTGGAAATTGATGACGAAGTATCACGACAGGAAGGAGTACATGGAACTGATGAAACAGTACAATCCAGATCAAGGGGCCCGTACCAATAAGCTGTACATAGATCCATGTGTGACTTTCAAGAATCCGTCTTATAGAGCGGATCCTATTACGGAAAGCTAG
Protein
MNSSDSLEDPLEPSWNLCPELRMENGCYTTFAYKYQDGYGIELLIQKNKNCDENILKLGVIVSVTVILVGIGTLVVWKLMTKYHDRKEYMELMKQYNPDQGARTNKLYIDPCVTFKNPSYRADPITES
Summary
Similarity
Belongs to the integrin beta chain family.
Feature
chain Integrin beta
Uniprot
A0A076JP59
A0A2H1V7X1
A0A2A4IXQ7
A0A212FB70
A0A194R3R5
A0A194PKP6
+ More
V9I2Y6 A0A3G5MWV8 A0A182HIV0 N6SYT4 A0A1B6JV91 A0A182XDN2 A0A182STS0 D6X0D6 A0A182M2H6 H9JBI3 S4PCV3 A0A076JVB8 D1MBK3 A0A2P2I7V9 A0A182KEI3 A0A1E1WNR2 A0A0P5WWD4 A0A2M4BIB1 A0A1Z5L8Z7 A0A1Y1KQM0 A0A0P5WC53 I4DPR3 A0A0P5WG12 A0A0K2UQU2 A0A2A4K6E0 A0A2M4BDW1 A0A2M4BDV9 A0A023UG44 A0A164YYE1 A0A2M4BIS4 A0A0N8B5P6 A0A0P5EN75 A0A194RCN7 A0A2M4BI31 A0A0P5IU01 A0A0P4Y6Q1 A0A0P6AIE1 E9GI17 A0A182PFW1 A0A0P5EIN5 A0A1Y1KLF1 U4USQ6 A0A1Y1KR32 D6WK71 A0A194PEH3 A2TIK8 A0A2M4ACS8 A0A2M4ACZ9 A0A0P5HS47 A0A2M4A9A0 A0A2M4A9N8 A0A182IJP6 A0A182YH54 A0A084VH17 T1GNV7 A0A0P4YNI8 A0A182RHJ6 A0A182W5K9 A0A212EPU0 X2BBV3 A0A2H1WGL4 A0A2M4CU05 F8TAD6 A0A182FIP7 A0A2M4CU18 A0A182Q0X5 T1DSJ7 A0A2M3ZGX5 A0A0G3BIA0 W8C804 W5JJT5 A0A3Q0JA47 A0A2P2I2S2 A0A034VPD2 A0A1I8PFU3 D1MCA9 A0A1S3IPR8 Q95P95 Q86G85 A0A210Q5V9 A0A076JQI1 D1MBK4 E2B5Z7 A0A0C9PS78 K9JDL3 P91774 B4Q0N6 A0A0R1EBH1 A0A336KMF2 E0W1M5 Q7QE55 A0A182V657 Q9NAS7
V9I2Y6 A0A3G5MWV8 A0A182HIV0 N6SYT4 A0A1B6JV91 A0A182XDN2 A0A182STS0 D6X0D6 A0A182M2H6 H9JBI3 S4PCV3 A0A076JVB8 D1MBK3 A0A2P2I7V9 A0A182KEI3 A0A1E1WNR2 A0A0P5WWD4 A0A2M4BIB1 A0A1Z5L8Z7 A0A1Y1KQM0 A0A0P5WC53 I4DPR3 A0A0P5WG12 A0A0K2UQU2 A0A2A4K6E0 A0A2M4BDW1 A0A2M4BDV9 A0A023UG44 A0A164YYE1 A0A2M4BIS4 A0A0N8B5P6 A0A0P5EN75 A0A194RCN7 A0A2M4BI31 A0A0P5IU01 A0A0P4Y6Q1 A0A0P6AIE1 E9GI17 A0A182PFW1 A0A0P5EIN5 A0A1Y1KLF1 U4USQ6 A0A1Y1KR32 D6WK71 A0A194PEH3 A2TIK8 A0A2M4ACS8 A0A2M4ACZ9 A0A0P5HS47 A0A2M4A9A0 A0A2M4A9N8 A0A182IJP6 A0A182YH54 A0A084VH17 T1GNV7 A0A0P4YNI8 A0A182RHJ6 A0A182W5K9 A0A212EPU0 X2BBV3 A0A2H1WGL4 A0A2M4CU05 F8TAD6 A0A182FIP7 A0A2M4CU18 A0A182Q0X5 T1DSJ7 A0A2M3ZGX5 A0A0G3BIA0 W8C804 W5JJT5 A0A3Q0JA47 A0A2P2I2S2 A0A034VPD2 A0A1I8PFU3 D1MCA9 A0A1S3IPR8 Q95P95 Q86G85 A0A210Q5V9 A0A076JQI1 D1MBK4 E2B5Z7 A0A0C9PS78 K9JDL3 P91774 B4Q0N6 A0A0R1EBH1 A0A336KMF2 E0W1M5 Q7QE55 A0A182V657 Q9NAS7
Pubmed
25064490
22118469
26354079
23537049
18362917
19820115
+ More
19121390 23622113 28528879 28004739 22651552 21292972 20708011 25244985 24438588 23254933 21856307 26004499 24495485 20920257 23761445 25348373 22792387 12697309 15990346 16442158 12974949 28812685 20798317 9062999 17994087 17550304 20566863 12364791 14747013 17210077 11437913
19121390 23622113 28528879 28004739 22651552 21292972 20708011 25244985 24438588 23254933 21856307 26004499 24495485 20920257 23761445 25348373 22792387 12697309 15990346 16442158 12974949 28812685 20798317 9062999 17994087 17550304 20566863 12364791 14747013 17210077 11437913
EMBL
KJ511856
AII79415.1
ODYU01001118
SOQ36886.1
NWSH01005801
PCG63940.1
+ More
AGBW02009374 OWR50969.1 KQ460855 KPJ11880.1 KQ459601 KPI94006.1 HQ260916 AEN94570.1 MH170220 AYX40719.1 APCN01004101 APGK01059069 APGK01059070 APGK01059071 APGK01059072 KB741292 ENN70393.1 GECU01004581 JAT03126.1 KQ971372 EFA10689.2 AXCM01014002 AXCM01014003 BABH01030933 BABH01030934 GAIX01005247 JAA87313.1 KJ511854 AII79413.1 GU129932 ACY74431.1 IACF01004524 LAB70113.1 GDQN01007634 GDQN01002429 JAT83420.1 JAT88625.1 GDIP01080635 JAM23080.1 GGFJ01003550 MBW52691.1 GFJQ02003302 JAW03668.1 GEZM01076841 JAV63604.1 GDIP01088141 JAM15574.1 AK403732 BAM19903.1 GDIP01087138 JAM16577.1 HACA01023064 CDW40425.1 NWSH01000130 PCG79230.1 GGFJ01002056 MBW51197.1 GGFJ01002095 MBW51236.1 KJ462516 AHY00655.1 LRGB01000868 KZS15734.1 GGFJ01003577 MBW52718.1 GDIQ01245981 GDIQ01227794 GDIQ01210291 GDIQ01207807 GDIQ01206458 GDIQ01203248 GDIQ01191690 GDIQ01132238 GDIQ01113476 GDIP01089236 JAK41434.1 JAM14479.1 GDIQ01269713 JAJ82011.1 KQ460398 KPJ15040.1 GGFJ01003578 MBW52719.1 GDIP01253007 GDIQ01269714 GDIQ01227795 GDIQ01227793 GDIQ01225644 GDIQ01210290 GDIQ01191691 JAI70394.1 JAK41435.1 GDIP01231880 JAI91521.1 GDIP01030205 JAM73510.1 GL732545 EFX80954.1 GDIQ01270948 JAJ80776.1 GEZM01080428 JAV62144.1 KB632349 ERL93211.1 GEZM01080427 JAV62145.1 KQ971342 EFA03618.2 KQ459606 KPI91428.1 EF202843 ABM91320.1 GGFK01005275 MBW38596.1 GGFK01005281 MBW38602.1 GDIQ01229031 JAK22694.1 GGFK01003991 MBW37312.1 GGFK01004007 MBW37328.1 ATLV01013123 KE524840 KFB37261.1 CAQQ02145466 CAQQ02145467 GDIP01224722 JAI98679.1 AGBW02013402 OWR43484.1 AMQN01000124 ODYU01008525 SOQ52209.1 GGFL01004658 MBW68836.1 HQ829425 ADW20295.1 GGFL01004659 MBW68837.1 AXCN02002227 GAMD01001449 JAB00142.1 GGFM01007066 MBW27817.1 KP795392 AKJ26284.1 GAMC01000222 JAC06334.1 ADMH02000934 ETN64637.1 IACF01002654 LAB68299.1 GAKP01014633 JAC44319.1 GU131148 ACY82398.1 AB066348 BAB62173.1 AY237588 AAO85806.1 NEDP02004918 OWF44108.1 KJ511855 AII79414.1 GU129933 ACY74432.1 GL445900 EFN88858.1 GBYB01004213 JAG73980.1 GQ889365 ADK56123.1 X98852 CAA67357.1 CM000162 EDX02307.1 KRK06701.1 UFQS01000673 UFQT01000673 SSX06026.1 SSX26383.1 DS235872 EEB19531.1 AAAB01008847 EAA06838.2 AJ292755 CAC00630.1
AGBW02009374 OWR50969.1 KQ460855 KPJ11880.1 KQ459601 KPI94006.1 HQ260916 AEN94570.1 MH170220 AYX40719.1 APCN01004101 APGK01059069 APGK01059070 APGK01059071 APGK01059072 KB741292 ENN70393.1 GECU01004581 JAT03126.1 KQ971372 EFA10689.2 AXCM01014002 AXCM01014003 BABH01030933 BABH01030934 GAIX01005247 JAA87313.1 KJ511854 AII79413.1 GU129932 ACY74431.1 IACF01004524 LAB70113.1 GDQN01007634 GDQN01002429 JAT83420.1 JAT88625.1 GDIP01080635 JAM23080.1 GGFJ01003550 MBW52691.1 GFJQ02003302 JAW03668.1 GEZM01076841 JAV63604.1 GDIP01088141 JAM15574.1 AK403732 BAM19903.1 GDIP01087138 JAM16577.1 HACA01023064 CDW40425.1 NWSH01000130 PCG79230.1 GGFJ01002056 MBW51197.1 GGFJ01002095 MBW51236.1 KJ462516 AHY00655.1 LRGB01000868 KZS15734.1 GGFJ01003577 MBW52718.1 GDIQ01245981 GDIQ01227794 GDIQ01210291 GDIQ01207807 GDIQ01206458 GDIQ01203248 GDIQ01191690 GDIQ01132238 GDIQ01113476 GDIP01089236 JAK41434.1 JAM14479.1 GDIQ01269713 JAJ82011.1 KQ460398 KPJ15040.1 GGFJ01003578 MBW52719.1 GDIP01253007 GDIQ01269714 GDIQ01227795 GDIQ01227793 GDIQ01225644 GDIQ01210290 GDIQ01191691 JAI70394.1 JAK41435.1 GDIP01231880 JAI91521.1 GDIP01030205 JAM73510.1 GL732545 EFX80954.1 GDIQ01270948 JAJ80776.1 GEZM01080428 JAV62144.1 KB632349 ERL93211.1 GEZM01080427 JAV62145.1 KQ971342 EFA03618.2 KQ459606 KPI91428.1 EF202843 ABM91320.1 GGFK01005275 MBW38596.1 GGFK01005281 MBW38602.1 GDIQ01229031 JAK22694.1 GGFK01003991 MBW37312.1 GGFK01004007 MBW37328.1 ATLV01013123 KE524840 KFB37261.1 CAQQ02145466 CAQQ02145467 GDIP01224722 JAI98679.1 AGBW02013402 OWR43484.1 AMQN01000124 ODYU01008525 SOQ52209.1 GGFL01004658 MBW68836.1 HQ829425 ADW20295.1 GGFL01004659 MBW68837.1 AXCN02002227 GAMD01001449 JAB00142.1 GGFM01007066 MBW27817.1 KP795392 AKJ26284.1 GAMC01000222 JAC06334.1 ADMH02000934 ETN64637.1 IACF01002654 LAB68299.1 GAKP01014633 JAC44319.1 GU131148 ACY82398.1 AB066348 BAB62173.1 AY237588 AAO85806.1 NEDP02004918 OWF44108.1 KJ511855 AII79414.1 GU129933 ACY74432.1 GL445900 EFN88858.1 GBYB01004213 JAG73980.1 GQ889365 ADK56123.1 X98852 CAA67357.1 CM000162 EDX02307.1 KRK06701.1 UFQS01000673 UFQT01000673 SSX06026.1 SSX26383.1 DS235872 EEB19531.1 AAAB01008847 EAA06838.2 AJ292755 CAC00630.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000075840
UP000019118
+ More
UP000076407 UP000075901 UP000007266 UP000075883 UP000005204 UP000075881 UP000076858 UP000000305 UP000075885 UP000030742 UP000075880 UP000076408 UP000030765 UP000015102 UP000075900 UP000075920 UP000069272 UP000075886 UP000000673 UP000079169 UP000095300 UP000085678 UP000242188 UP000008237 UP000002282 UP000009046 UP000007062 UP000075903
UP000076407 UP000075901 UP000007266 UP000075883 UP000005204 UP000075881 UP000076858 UP000000305 UP000075885 UP000030742 UP000075880 UP000076408 UP000030765 UP000015102 UP000075900 UP000075920 UP000069272 UP000075886 UP000000673 UP000079169 UP000095300 UP000085678 UP000242188 UP000008237 UP000002282 UP000009046 UP000007062 UP000075903
Pfam
Interpro
IPR015812
Integrin_bsu
+ More
IPR036465 vWFA_dom_sf
IPR002369 Integrin_bsu_VWA
IPR002035 VWF_A
IPR013111 EGF_extracell
IPR014836 Integrin_bsu_cyt_dom
IPR040622 I-EGF_1
IPR036349 Integrin_bsu_tail_dom_sf
IPR032695 Integrin_dom_sf
IPR012896 Integrin_bsu_tail
IPR033760 Integrin_beta_N
IPR027071 Integrin_beta-1
IPR027068 Integrin_beta-3
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR036465 vWFA_dom_sf
IPR002369 Integrin_bsu_VWA
IPR002035 VWF_A
IPR013111 EGF_extracell
IPR014836 Integrin_bsu_cyt_dom
IPR040622 I-EGF_1
IPR036349 Integrin_bsu_tail_dom_sf
IPR032695 Integrin_dom_sf
IPR012896 Integrin_bsu_tail
IPR033760 Integrin_beta_N
IPR027071 Integrin_beta-1
IPR027068 Integrin_beta-3
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
Gene 3D
ProteinModelPortal
A0A076JP59
A0A2H1V7X1
A0A2A4IXQ7
A0A212FB70
A0A194R3R5
A0A194PKP6
+ More
V9I2Y6 A0A3G5MWV8 A0A182HIV0 N6SYT4 A0A1B6JV91 A0A182XDN2 A0A182STS0 D6X0D6 A0A182M2H6 H9JBI3 S4PCV3 A0A076JVB8 D1MBK3 A0A2P2I7V9 A0A182KEI3 A0A1E1WNR2 A0A0P5WWD4 A0A2M4BIB1 A0A1Z5L8Z7 A0A1Y1KQM0 A0A0P5WC53 I4DPR3 A0A0P5WG12 A0A0K2UQU2 A0A2A4K6E0 A0A2M4BDW1 A0A2M4BDV9 A0A023UG44 A0A164YYE1 A0A2M4BIS4 A0A0N8B5P6 A0A0P5EN75 A0A194RCN7 A0A2M4BI31 A0A0P5IU01 A0A0P4Y6Q1 A0A0P6AIE1 E9GI17 A0A182PFW1 A0A0P5EIN5 A0A1Y1KLF1 U4USQ6 A0A1Y1KR32 D6WK71 A0A194PEH3 A2TIK8 A0A2M4ACS8 A0A2M4ACZ9 A0A0P5HS47 A0A2M4A9A0 A0A2M4A9N8 A0A182IJP6 A0A182YH54 A0A084VH17 T1GNV7 A0A0P4YNI8 A0A182RHJ6 A0A182W5K9 A0A212EPU0 X2BBV3 A0A2H1WGL4 A0A2M4CU05 F8TAD6 A0A182FIP7 A0A2M4CU18 A0A182Q0X5 T1DSJ7 A0A2M3ZGX5 A0A0G3BIA0 W8C804 W5JJT5 A0A3Q0JA47 A0A2P2I2S2 A0A034VPD2 A0A1I8PFU3 D1MCA9 A0A1S3IPR8 Q95P95 Q86G85 A0A210Q5V9 A0A076JQI1 D1MBK4 E2B5Z7 A0A0C9PS78 K9JDL3 P91774 B4Q0N6 A0A0R1EBH1 A0A336KMF2 E0W1M5 Q7QE55 A0A182V657 Q9NAS7
V9I2Y6 A0A3G5MWV8 A0A182HIV0 N6SYT4 A0A1B6JV91 A0A182XDN2 A0A182STS0 D6X0D6 A0A182M2H6 H9JBI3 S4PCV3 A0A076JVB8 D1MBK3 A0A2P2I7V9 A0A182KEI3 A0A1E1WNR2 A0A0P5WWD4 A0A2M4BIB1 A0A1Z5L8Z7 A0A1Y1KQM0 A0A0P5WC53 I4DPR3 A0A0P5WG12 A0A0K2UQU2 A0A2A4K6E0 A0A2M4BDW1 A0A2M4BDV9 A0A023UG44 A0A164YYE1 A0A2M4BIS4 A0A0N8B5P6 A0A0P5EN75 A0A194RCN7 A0A2M4BI31 A0A0P5IU01 A0A0P4Y6Q1 A0A0P6AIE1 E9GI17 A0A182PFW1 A0A0P5EIN5 A0A1Y1KLF1 U4USQ6 A0A1Y1KR32 D6WK71 A0A194PEH3 A2TIK8 A0A2M4ACS8 A0A2M4ACZ9 A0A0P5HS47 A0A2M4A9A0 A0A2M4A9N8 A0A182IJP6 A0A182YH54 A0A084VH17 T1GNV7 A0A0P4YNI8 A0A182RHJ6 A0A182W5K9 A0A212EPU0 X2BBV3 A0A2H1WGL4 A0A2M4CU05 F8TAD6 A0A182FIP7 A0A2M4CU18 A0A182Q0X5 T1DSJ7 A0A2M3ZGX5 A0A0G3BIA0 W8C804 W5JJT5 A0A3Q0JA47 A0A2P2I2S2 A0A034VPD2 A0A1I8PFU3 D1MCA9 A0A1S3IPR8 Q95P95 Q86G85 A0A210Q5V9 A0A076JQI1 D1MBK4 E2B5Z7 A0A0C9PS78 K9JDL3 P91774 B4Q0N6 A0A0R1EBH1 A0A336KMF2 E0W1M5 Q7QE55 A0A182V657 Q9NAS7
PDB
4UM8
E-value=7.1772e-07,
Score=120
Ontologies
GO
GO:0007155
GO:0007229
GO:0016021
GO:0008305
GO:0038023
GO:0007160
GO:0070527
GO:0005178
GO:0009986
GO:0033627
GO:0005925
GO:0016477
GO:0034446
GO:0042734
GO:0045211
GO:0008360
GO:0030336
GO:0048803
GO:0043034
GO:0045214
GO:0030425
GO:0007608
GO:0007419
GO:0090129
GO:0007411
GO:0030718
GO:0005927
GO:0009925
GO:0008340
GO:0007391
GO:0035001
GO:0051492
GO:0046982
GO:0006930
GO:0003344
GO:1990430
GO:0007431
GO:0007377
GO:0016203
GO:0035099
GO:0016328
GO:0007298
GO:0021551
GO:0007015
GO:0035160
GO:0007508
GO:0031252
PANTHER
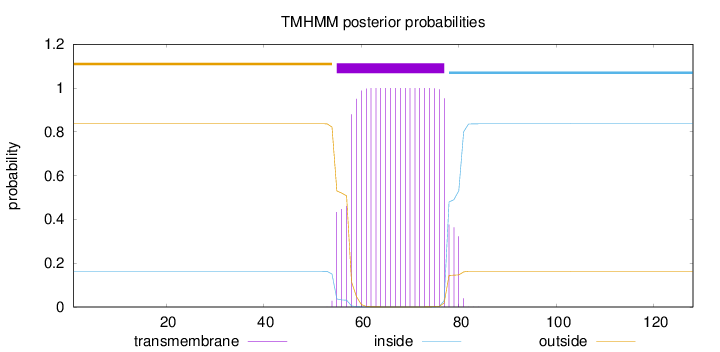
Topology
Subcellular location
Membrane
Length:
128
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.23468
Exp number, first 60 AAs:
4.19147
Total prob of N-in:
0.16302
outside
1 - 54
TMhelix
55 - 77
inside
78 - 128
Population Genetic Test Statistics
Pi
212.910299
Theta
20.876249
Tajima's D
0.710738
CLR
0.47998
CSRT
0.587820608969552
Interpretation
Uncertain
