Gene
KWMTBOMO01778
Pre Gene Modal
BGIBMGA006002
Annotation
integrin_beta2_[Bombyx_mori]
Full name
Integrin beta
Location in the cell
PlasmaMembrane Reliability : 4.894
Sequence
CDS
ATGCTTTATAAACCTATAGAGCATTTCCCGCTCGATGTTTATTTTCTCATGGATAACTCTTATACTATGAGACAATTTCAGAACGAACTAAAATCACAGGCTATTAATATTTTGAAAGAATTAAGTGCTTTCACAAAGAATGTTCGATTGGGATTCGGAACCTTTGTAGAAAAACCAGTTTATCCATATTACGATAAAAACAGATACCAAAAGTCCATACCGTTTGAAAATGTATTATCCTTAACGGCAGACATATCGAAATTGAACAACACAGTTCGCCAAATCGATTTTGGTTCAAATTTCGATGACCAAGAAGCTGGCTTAGATGCTCTGATGCAGGTTATGACCTGCACCAAAGAAATAGGGTGGCGAACAGAAGCAAGACGTATTATAGTACTCTTCACTGACGCGCCGTATCATAGTATGGGAGACGGTAAAATGATCGGCATATTAAAACCGAATGATATGGAATGCCACCTAAAAGATAATATTTATGACGATACAGCAACGACACAAGATTACCCGTCTGTAAGCCAGATAAATAAAGTAGCAACAGAAAGAAATTTCAAAATCATATTTGCTACTATTGAAAGGGTATCCGAGGTTTACAAAGAATTGTCAAATGCAATACTAGGATCAGAAACAGCAATTTTAATTAAAAAAGAGGAAGGGGAGTCAAATATTGTGCCTATAATTAAAACTGCCTATTTGGAAGCCGAAAGTGTTGTAAGGCTAGTATCTGATTTTCCAAGTTCAGTTAATGTCAAGTATAGTCCGGATTGCAATGAAATTGAAAAGAAAAATTGCAGAAGTACACATTTAAATCCCATTGTAGAAATAAATGTTACAATAGTTGCTGGATCGTGTCTCGATGGAAATTCTTCTAGAGTCTTTAAAATAAGTCCGCATGGTCTTCATGATATTGACGTAGGTGTTGAGTTAGAGGTTGACTGTCAATGTGAATGCGAGAAGGTAAAAAAAGAGAAGAGTCCCGTTTGTTTTAATGCTGCGTATTTATTGTGCGGTGTATGCATCTGTGACAGTGACAGTAAGGGATCAAACGTGGCCGAAGATTTGGAAAGCCCTCACTGTAGATATCATGGACAATACGTTTGCGGCCGTTGTATTTGTAAGCCCGGATATTCAGGTGACACGTGCGAATATGATGATAATTTGTGTATGAGTAACTGCAAAAAGGGAACATGCTATATGGGTAAATGTAATTGTCAAGAGGGCTGGTCGCCTCCGAACTGTATGTGTCCTGTAAGCAACAAAGATTGCATAGCGCCGTGGTCAGACGAGAATGAGGTTTGCTTCGGCAACGGGGAATGTAAATGTGGAGAATGTTCCTGTGATAAAGTTAGCGGCAAAAATGAATCGTATTTCGGCACATTCTGCGACTTATGCACTGATTGCGCCGCAAAATACTGTAAAAAATATGAAGACTACGTATATTGTAATTACTTGAACGAAAAGGCGTTTTGTGATCAATCATTCTTGAACGGAAAGGACACGGTTGTTAAGATTGTTAATTCAACGGAAATCATAAATGATAACATTCAAAAGGCTACTTGGTGTGCCAAGCTATTGGATAACGGAACATCGATTGTGTTCAAATACCACCACGAGGACAATAAACTTTTTTTGACTATACAAAAGGAAAAGCAGATACCGCCTAAAGCTAATTTATATATTGTTGTGGGTAGTGTGATCTCTGCTATTGTATTGATTGGCTTGCTGACTCTCATCGGATGGAAGGTTTTAACAGATATCTACGATCAAAGGGAATACAGGAAAGTGGAAGAAGCGGCCAAAGCCGCCGGCTTTGATGTTTCAAATCCTTGTTACGAAAACCCGACTACAGAATTTCCGAACCCATCTTACAGACCTACGAAATAA
Protein
MLYKPIEHFPLDVYFLMDNSYTMRQFQNELKSQAINILKELSAFTKNVRLGFGTFVEKPVYPYYDKNRYQKSIPFENVLSLTADISKLNNTVRQIDFGSNFDDQEAGLDALMQVMTCTKEIGWRTEARRIIVLFTDAPYHSMGDGKMIGILKPNDMECHLKDNIYDDTATTQDYPSVSQINKVATERNFKIIFATIERVSEVYKELSNAILGSETAILIKKEEGESNIVPIIKTAYLEAESVVRLVSDFPSSVNVKYSPDCNEIEKKNCRSTHLNPIVEINVTIVAGSCLDGNSSRVFKISPHGLHDIDVGVELEVDCQCECEKVKKEKSPVCFNAAYLLCGVCICDSDSKGSNVAEDLESPHCRYHGQYVCGRCICKPGYSGDTCEYDDNLCMSNCKKGTCYMGKCNCQEGWSPPNCMCPVSNKDCIAPWSDENEVCFGNGECKCGECSCDKVSGKNESYFGTFCDLCTDCAAKYCKKYEDYVYCNYLNEKAFCDQSFLNGKDTVVKIVNSTEIINDNIQKATWCAKLLDNGTSIVFKYHHEDNKLFLTIQKEKQIPPKANLYIVVGSVISAIVLIGLLTLIGWKVLTDIYDQREYRKVEEAAKAAGFDVSNPCYENPTTEFPNPSYRPTK
Summary
Similarity
Belongs to the integrin beta chain family.
Feature
chain Integrin beta
Uniprot
A0A076JQI1
D1MBK4
H9J910
Q66PY4
A0A2A4IWA0
A0S6A3
+ More
A0A194PMP9 A0A212FB66 A0A194R7M9 A0A0L7KWJ3 E0D5F3 A0A158NE45 E9JBK8 A0A195BPC3 E2A0M9 F4W856 A0A195EYJ7 A0A195D7K9 A0A0J7L3M3 A0A151X462 Q95P95 A0A084VH17 K7IP94 A0A1B6GQB5 A0A3Q0JA47 A0A1S3I615 A0A1S3JZW7 A0A2A3ECL4 A0A182PFW1 A0A182IJP6 A0A182XDN2 A0A2A4K6E0 A0A182HIV0 A0A087ZTR1 A0A067QK14 A0A154PGR4 A0A232F1V4 H9JBI3 A0A023UG44 A0A076JVB8 D1MBK3 A0A336KRK3 A0A0P5EN75 A0A182W5K9 A0A1W4WXC9 A0A182YH54 V9I2Y6 A0A182RHJ6 A0A0N8B5P6 A0A1Y1KQM0 A0A2M4BDV9 A0A2M4BDW1 A0A0P5IU01 A0A336MVT2 A0A182Q0X5 A0A2M4CU18 A0A2M4CU05 C6KE04 A0A1S3I8E3 D6X0D6 A0A3L8DYQ0 A0A026W443 A0A1Z5L7T6 A0A2H1WGL4 F8TAD6 D6WK71 A0A1E1WNR2 E9GI17 A0A164YYE1 A0A2D3E2V7 A0A212EPU0 A0A3P6TGD1 A0A1A9X588 A0A0G3BIA0 A0A336M8L1 A0A1A9UUR6 A0A0K8UTI8 A0A182TIH0 A0A1A9X8U0 A0A1B0AX62 B4JNC9 A0A1S3IPR8 A0A1I8PFU3 A0A336MBS4 A0A182V657 Q7QE55 Q16M01 Q86G85 A0A1S4FW80 X2BBV3 A0A034V7S9 A0A0P6IR12 A0A0A1WKS8 Q9NAS7 E0VEZ0 A0A2A3E010 L7M1I1 A0A1B6MUI8 A0A069DXI6 A0A1I8PZ80
A0A194PMP9 A0A212FB66 A0A194R7M9 A0A0L7KWJ3 E0D5F3 A0A158NE45 E9JBK8 A0A195BPC3 E2A0M9 F4W856 A0A195EYJ7 A0A195D7K9 A0A0J7L3M3 A0A151X462 Q95P95 A0A084VH17 K7IP94 A0A1B6GQB5 A0A3Q0JA47 A0A1S3I615 A0A1S3JZW7 A0A2A3ECL4 A0A182PFW1 A0A182IJP6 A0A182XDN2 A0A2A4K6E0 A0A182HIV0 A0A087ZTR1 A0A067QK14 A0A154PGR4 A0A232F1V4 H9JBI3 A0A023UG44 A0A076JVB8 D1MBK3 A0A336KRK3 A0A0P5EN75 A0A182W5K9 A0A1W4WXC9 A0A182YH54 V9I2Y6 A0A182RHJ6 A0A0N8B5P6 A0A1Y1KQM0 A0A2M4BDV9 A0A2M4BDW1 A0A0P5IU01 A0A336MVT2 A0A182Q0X5 A0A2M4CU18 A0A2M4CU05 C6KE04 A0A1S3I8E3 D6X0D6 A0A3L8DYQ0 A0A026W443 A0A1Z5L7T6 A0A2H1WGL4 F8TAD6 D6WK71 A0A1E1WNR2 E9GI17 A0A164YYE1 A0A2D3E2V7 A0A212EPU0 A0A3P6TGD1 A0A1A9X588 A0A0G3BIA0 A0A336M8L1 A0A1A9UUR6 A0A0K8UTI8 A0A182TIH0 A0A1A9X8U0 A0A1B0AX62 B4JNC9 A0A1S3IPR8 A0A1I8PFU3 A0A336MBS4 A0A182V657 Q7QE55 Q16M01 Q86G85 A0A1S4FW80 X2BBV3 A0A034V7S9 A0A0P6IR12 A0A0A1WKS8 Q9NAS7 E0VEZ0 A0A2A3E010 L7M1I1 A0A1B6MUI8 A0A069DXI6 A0A1I8PZ80
Pubmed
25064490
19121390
15804572
26354079
22118469
26227816
+ More
21347285 21282665 20798317 21719571 12697309 15990346 16442158 24438588 20075255 24845553 28648823 25244985 28004739 18362917 19820115 30249741 24508170 28528879 21856307 21292972 26004499 17994087 12364791 14747013 17210077 17510324 12974949 23254933 25348373 25830018 11437913 20566863 25576852 26334808
21347285 21282665 20798317 21719571 12697309 15990346 16442158 24438588 20075255 24845553 28648823 25244985 28004739 18362917 19820115 30249741 24508170 28528879 21856307 21292972 26004499 17994087 12364791 14747013 17210077 17510324 12974949 23254933 25348373 25830018 11437913 20566863 25576852 26334808
EMBL
KJ511855
AII79414.1
GU129933
ACY74432.1
BABH01004742
BABH01004743
+ More
AY630342 AAU11316.1 NWSH01005801 PCG63939.1 DQ289584 ABB92837.1 KQ459601 KPI94009.1 AGBW02009374 OWR50970.1 KQ460855 KPJ11876.1 JTDY01005041 KOB67436.1 AB548819 BAJ16205.1 ADTU01012794 ADTU01012795 GL771210 EFZ09795.1 KQ976424 KYM88360.1 GL435626 EFN72932.1 GL887888 EGI69726.1 KQ981906 KYN33293.1 KQ981153 KYN08861.1 LBMM01001036 KMQ97039.1 KQ982548 KYQ55161.1 AB066348 BAB62173.1 ATLV01013123 KE524840 KFB37261.1 GECZ01005141 JAS64628.1 KZ288291 PBC29224.1 NWSH01000130 PCG79230.1 APCN01004101 KK853366 KDR08103.1 KQ434899 KZC10997.1 NNAY01001290 OXU24469.1 BABH01030933 BABH01030934 KJ462516 AHY00655.1 KJ511854 AII79413.1 GU129932 ACY74431.1 UFQS01000673 UFQT01000673 SSX06025.1 SSX26382.1 GDIQ01269713 JAJ82011.1 HQ260916 AEN94570.1 GDIQ01245981 GDIQ01227794 GDIQ01210291 GDIQ01207807 GDIQ01206458 GDIQ01203248 GDIQ01191690 GDIQ01132238 GDIQ01113476 GDIP01089236 JAK41434.1 JAM14479.1 GEZM01076841 JAV63604.1 GGFJ01002095 MBW51236.1 GGFJ01002056 MBW51197.1 GDIP01253007 GDIQ01269714 GDIQ01227795 GDIQ01227793 GDIQ01225644 GDIQ01210290 GDIQ01191691 JAI70394.1 JAK41435.1 UFQS01003187 UFQT01003187 SSX15340.1 SSX34714.1 AXCN02002227 GGFL01004659 MBW68837.1 GGFL01004658 MBW68836.1 GQ178290 ACS66819.1 KQ971372 EFA10689.2 QOIP01000002 RLU25353.1 KK107436 EZA50855.1 GFJQ02003483 JAW03487.1 ODYU01008525 SOQ52209.1 HQ829425 ADW20295.1 KQ971342 EFA03618.2 GDQN01007634 GDQN01002429 JAT83420.1 JAT88625.1 GL732545 EFX80954.1 LRGB01000868 KZS15734.1 MF140473 ATU31738.1 AGBW02013402 OWR43484.1 UYRX01000315 VDK79935.1 KP795392 AKJ26284.1 SSX26380.1 GDHF01022317 JAI29997.1 JXJN01005087 JXJN01005088 CH916371 EDV92222.1 SSX26379.1 AAAB01008847 EAA06838.2 CH477885 EAT35362.1 AY237588 AAO85806.1 AMQN01000124 GAKP01019606 JAC39346.1 GDIQ01007080 JAN87657.1 GBXI01015167 JAC99124.1 AJ292755 CAC00630.1 DS235100 EEB11964.1 KZ288518 PBC25100.1 GACK01008080 JAA56954.1 GEBQ01000406 JAT39571.1 GBGD01000492 JAC88397.1
AY630342 AAU11316.1 NWSH01005801 PCG63939.1 DQ289584 ABB92837.1 KQ459601 KPI94009.1 AGBW02009374 OWR50970.1 KQ460855 KPJ11876.1 JTDY01005041 KOB67436.1 AB548819 BAJ16205.1 ADTU01012794 ADTU01012795 GL771210 EFZ09795.1 KQ976424 KYM88360.1 GL435626 EFN72932.1 GL887888 EGI69726.1 KQ981906 KYN33293.1 KQ981153 KYN08861.1 LBMM01001036 KMQ97039.1 KQ982548 KYQ55161.1 AB066348 BAB62173.1 ATLV01013123 KE524840 KFB37261.1 GECZ01005141 JAS64628.1 KZ288291 PBC29224.1 NWSH01000130 PCG79230.1 APCN01004101 KK853366 KDR08103.1 KQ434899 KZC10997.1 NNAY01001290 OXU24469.1 BABH01030933 BABH01030934 KJ462516 AHY00655.1 KJ511854 AII79413.1 GU129932 ACY74431.1 UFQS01000673 UFQT01000673 SSX06025.1 SSX26382.1 GDIQ01269713 JAJ82011.1 HQ260916 AEN94570.1 GDIQ01245981 GDIQ01227794 GDIQ01210291 GDIQ01207807 GDIQ01206458 GDIQ01203248 GDIQ01191690 GDIQ01132238 GDIQ01113476 GDIP01089236 JAK41434.1 JAM14479.1 GEZM01076841 JAV63604.1 GGFJ01002095 MBW51236.1 GGFJ01002056 MBW51197.1 GDIP01253007 GDIQ01269714 GDIQ01227795 GDIQ01227793 GDIQ01225644 GDIQ01210290 GDIQ01191691 JAI70394.1 JAK41435.1 UFQS01003187 UFQT01003187 SSX15340.1 SSX34714.1 AXCN02002227 GGFL01004659 MBW68837.1 GGFL01004658 MBW68836.1 GQ178290 ACS66819.1 KQ971372 EFA10689.2 QOIP01000002 RLU25353.1 KK107436 EZA50855.1 GFJQ02003483 JAW03487.1 ODYU01008525 SOQ52209.1 HQ829425 ADW20295.1 KQ971342 EFA03618.2 GDQN01007634 GDQN01002429 JAT83420.1 JAT88625.1 GL732545 EFX80954.1 LRGB01000868 KZS15734.1 MF140473 ATU31738.1 AGBW02013402 OWR43484.1 UYRX01000315 VDK79935.1 KP795392 AKJ26284.1 SSX26380.1 GDHF01022317 JAI29997.1 JXJN01005087 JXJN01005088 CH916371 EDV92222.1 SSX26379.1 AAAB01008847 EAA06838.2 CH477885 EAT35362.1 AY237588 AAO85806.1 AMQN01000124 GAKP01019606 JAC39346.1 GDIQ01007080 JAN87657.1 GBXI01015167 JAC99124.1 AJ292755 CAC00630.1 DS235100 EEB11964.1 KZ288518 PBC25100.1 GACK01008080 JAA56954.1 GEBQ01000406 JAT39571.1 GBGD01000492 JAC88397.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000005205 UP000078540 UP000000311 UP000007755 UP000078541 UP000078492 UP000036403 UP000075809 UP000030765 UP000002358 UP000079169 UP000085678 UP000242457 UP000075885 UP000075880 UP000076407 UP000075840 UP000005203 UP000027135 UP000076502 UP000215335 UP000075920 UP000192223 UP000076408 UP000075900 UP000075886 UP000007266 UP000279307 UP000053097 UP000000305 UP000076858 UP000277928 UP000091820 UP000078200 UP000075902 UP000092443 UP000092460 UP000001070 UP000095300 UP000075903 UP000007062 UP000008820 UP000009046
UP000005205 UP000078540 UP000000311 UP000007755 UP000078541 UP000078492 UP000036403 UP000075809 UP000030765 UP000002358 UP000079169 UP000085678 UP000242457 UP000075885 UP000075880 UP000076407 UP000075840 UP000005203 UP000027135 UP000076502 UP000215335 UP000075920 UP000192223 UP000076408 UP000075900 UP000075886 UP000007266 UP000279307 UP000053097 UP000000305 UP000076858 UP000277928 UP000091820 UP000078200 UP000075902 UP000092443 UP000092460 UP000001070 UP000095300 UP000075903 UP000007062 UP000008820 UP000009046
Pfam
Interpro
IPR002369
Integrin_bsu_VWA
+ More
IPR002035 VWF_A
IPR015812 Integrin_bsu
IPR014836 Integrin_bsu_cyt_dom
IPR036465 vWFA_dom_sf
IPR040622 I-EGF_1
IPR032695 Integrin_dom_sf
IPR013032 EGF-like_CS
IPR013111 EGF_extracell
IPR036349 Integrin_bsu_tail_dom_sf
IPR012896 Integrin_bsu_tail
IPR033760 Integrin_beta_N
IPR000742 EGF-like_dom
IPR016201 PSI
IPR002035 VWF_A
IPR015812 Integrin_bsu
IPR014836 Integrin_bsu_cyt_dom
IPR036465 vWFA_dom_sf
IPR040622 I-EGF_1
IPR032695 Integrin_dom_sf
IPR013032 EGF-like_CS
IPR013111 EGF_extracell
IPR036349 Integrin_bsu_tail_dom_sf
IPR012896 Integrin_bsu_tail
IPR033760 Integrin_beta_N
IPR000742 EGF-like_dom
IPR016201 PSI
Gene 3D
ProteinModelPortal
A0A076JQI1
D1MBK4
H9J910
Q66PY4
A0A2A4IWA0
A0S6A3
+ More
A0A194PMP9 A0A212FB66 A0A194R7M9 A0A0L7KWJ3 E0D5F3 A0A158NE45 E9JBK8 A0A195BPC3 E2A0M9 F4W856 A0A195EYJ7 A0A195D7K9 A0A0J7L3M3 A0A151X462 Q95P95 A0A084VH17 K7IP94 A0A1B6GQB5 A0A3Q0JA47 A0A1S3I615 A0A1S3JZW7 A0A2A3ECL4 A0A182PFW1 A0A182IJP6 A0A182XDN2 A0A2A4K6E0 A0A182HIV0 A0A087ZTR1 A0A067QK14 A0A154PGR4 A0A232F1V4 H9JBI3 A0A023UG44 A0A076JVB8 D1MBK3 A0A336KRK3 A0A0P5EN75 A0A182W5K9 A0A1W4WXC9 A0A182YH54 V9I2Y6 A0A182RHJ6 A0A0N8B5P6 A0A1Y1KQM0 A0A2M4BDV9 A0A2M4BDW1 A0A0P5IU01 A0A336MVT2 A0A182Q0X5 A0A2M4CU18 A0A2M4CU05 C6KE04 A0A1S3I8E3 D6X0D6 A0A3L8DYQ0 A0A026W443 A0A1Z5L7T6 A0A2H1WGL4 F8TAD6 D6WK71 A0A1E1WNR2 E9GI17 A0A164YYE1 A0A2D3E2V7 A0A212EPU0 A0A3P6TGD1 A0A1A9X588 A0A0G3BIA0 A0A336M8L1 A0A1A9UUR6 A0A0K8UTI8 A0A182TIH0 A0A1A9X8U0 A0A1B0AX62 B4JNC9 A0A1S3IPR8 A0A1I8PFU3 A0A336MBS4 A0A182V657 Q7QE55 Q16M01 Q86G85 A0A1S4FW80 X2BBV3 A0A034V7S9 A0A0P6IR12 A0A0A1WKS8 Q9NAS7 E0VEZ0 A0A2A3E010 L7M1I1 A0A1B6MUI8 A0A069DXI6 A0A1I8PZ80
A0A194PMP9 A0A212FB66 A0A194R7M9 A0A0L7KWJ3 E0D5F3 A0A158NE45 E9JBK8 A0A195BPC3 E2A0M9 F4W856 A0A195EYJ7 A0A195D7K9 A0A0J7L3M3 A0A151X462 Q95P95 A0A084VH17 K7IP94 A0A1B6GQB5 A0A3Q0JA47 A0A1S3I615 A0A1S3JZW7 A0A2A3ECL4 A0A182PFW1 A0A182IJP6 A0A182XDN2 A0A2A4K6E0 A0A182HIV0 A0A087ZTR1 A0A067QK14 A0A154PGR4 A0A232F1V4 H9JBI3 A0A023UG44 A0A076JVB8 D1MBK3 A0A336KRK3 A0A0P5EN75 A0A182W5K9 A0A1W4WXC9 A0A182YH54 V9I2Y6 A0A182RHJ6 A0A0N8B5P6 A0A1Y1KQM0 A0A2M4BDV9 A0A2M4BDW1 A0A0P5IU01 A0A336MVT2 A0A182Q0X5 A0A2M4CU18 A0A2M4CU05 C6KE04 A0A1S3I8E3 D6X0D6 A0A3L8DYQ0 A0A026W443 A0A1Z5L7T6 A0A2H1WGL4 F8TAD6 D6WK71 A0A1E1WNR2 E9GI17 A0A164YYE1 A0A2D3E2V7 A0A212EPU0 A0A3P6TGD1 A0A1A9X588 A0A0G3BIA0 A0A336M8L1 A0A1A9UUR6 A0A0K8UTI8 A0A182TIH0 A0A1A9X8U0 A0A1B0AX62 B4JNC9 A0A1S3IPR8 A0A1I8PFU3 A0A336MBS4 A0A182V657 Q7QE55 Q16M01 Q86G85 A0A1S4FW80 X2BBV3 A0A034V7S9 A0A0P6IR12 A0A0A1WKS8 Q9NAS7 E0VEZ0 A0A2A3E010 L7M1I1 A0A1B6MUI8 A0A069DXI6 A0A1I8PZ80
PDB
4UM8
E-value=2.42756e-60,
Score=590
Ontologies
PATHWAY
GO
PANTHER
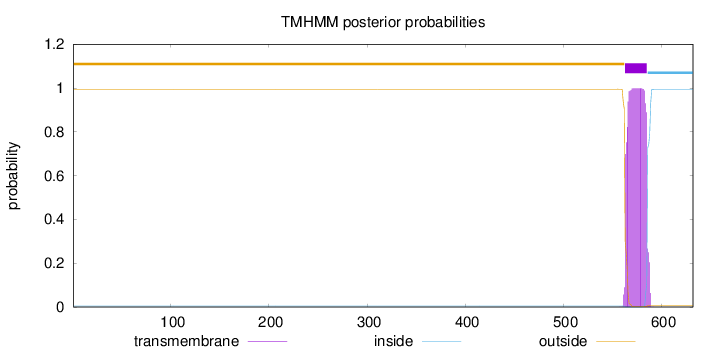
Topology
Subcellular location
Membrane
Length:
632
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.88968
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.00508
outside
1 - 562
TMhelix
563 - 585
inside
586 - 632
Population Genetic Test Statistics
Pi
201.085915
Theta
167.793525
Tajima's D
0.739488
CLR
0.283247
CSRT
0.582320883955802
Interpretation
Uncertain
