Gene
KWMTBOMO01776
Pre Gene Modal
BGIBMGA006149
Annotation
PREDICTED:_lipase_3-like_[Amyelois_transitella]
Full name
Lipase
Location in the cell
Mitochondrial Reliability : 1.656
Sequence
CDS
ATGTTTAGATATTTTATAACGACAATCTTAGTTTTCTCTTCTAATTACTGCTATGCAGAAATATCAGATGAAATCTTTTATAGAAATATTCAGAGGATCACTGATCCAAGTATACTGGAAGATGCGCGTTTAGATACGTTCACATTAATCCGTAAATACGGCTACCCGTGCGAAATACACAGAGTATACACAGAAGATGGATACATATTGGAAATGCATAGAGTACCTAACGGCAAATACAACGCGAAAAAAGGCCCCAAGCCGGTTGTGCTCCTACAGCATGGACTACTGTCGTCCTCTGCGGAATGGGTGCTTATGACCCCCGGGAAAGGTTTAGCATACATCTTAGCTGAAATCGGATATGACGTCTGGATGGGAAACGCTAGAGGAAACACGTATTCTCGTCATCACAAATCCTTAGTGGCCACTTCCTCGGAGTTTTGGAAATTCAGCTGGCACGAAATAGGTTACTACGACCTGCCCGCAATGATAGACTACGTCATCAAAGAAACCGGTGCACCTAAAATTCAGTATATTGGATTCTCTCAGGGCACCACAGTGTTCTGGGTGATGACGTCCACTCGGCCGGAATACAACGACAAGGTTTCGGCTATGCAAGCTTTAGCCCCGGTCGTGTATCTACGAAACGTGAGGAGCCCTCTTGTGAAGGCCATTGCTCCATTTACGAACACTTTGGAAATAATATTCAAGTTATTAGGAAAAAACGAGCTATTACCGAACGGCAAAATTAACGAACTTGCCGGACAAACTGTATGTGTTGAGGAAGCTATCACGCAGTTCCTTTGTACGAATCTACTGTTCCTAGTCTGTGGTTTCAACGAGGAGCAATTGAATAAAACAATGCTGCCAGTAGTGATGGGTCACACTCCAGCGGGCTCATCCACAAGGCAATTCGTACATTTCGGTCAGATTTACAAATCCGATGAGTTCTTACAATTCAATCACGGGTGGATAAAGAACAAATTCGTTTACGGTACATTTAGACCGCCCGCATACAATCTGAGCGCGATTCGGACACCTGCATTCCTTCACTACGCTGACAACGACTGGTTGTCGACACCGAATGACGTCCATAGACTATCAAGATCCTTGAAATCCGTCGTCGGCAAGTTCAAAGTACCCATGCCAGAATTCAATCATTTAGATTTCGTTTTTGCCATCAACGCAACGAATTACATATACTCTAGATTAATAAACATAATGGAACAGTTCAAGTAG
Protein
MFRYFITTILVFSSNYCYAEISDEIFYRNIQRITDPSILEDARLDTFTLIRKYGYPCEIHRVYTEDGYILEMHRVPNGKYNAKKGPKPVVLLQHGLLSSSAEWVLMTPGKGLAYILAEIGYDVWMGNARGNTYSRHHKSLVATSSEFWKFSWHEIGYYDLPAMIDYVIKETGAPKIQYIGFSQGTTVFWVMTSTRPEYNDKVSAMQALAPVVYLRNVRSPLVKAIAPFTNTLEIIFKLLGKNELLPNGKINELAGQTVCVEEAITQFLCTNLLFLVCGFNEEQLNKTMLPVVMGHTPAGSSTRQFVHFGQIYKSDEFLQFNHGWIKNKFVYGTFRPPAYNLSAIRTPAFLHYADNDWLSTPNDVHRLSRSLKSVVGKFKVPMPEFNHLDFVFAINATNYIYSRLINIMEQFK
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
A0A2A4IVE2
A0A2H1X0E5
A0A212FB51
A0A194R2Y3
A0A194PL00
H9J9F6
+ More
A0A1A9WZU7 A0A1A9UTK2 H9JQH1 A0A2A4K5R5 W8C916 A0A1L8DQ21 A0A1L8EIM3 A0A1L8EII7 A0A1L8EIH0 A0A1L8EID0 A0A2A4K5V7 A0A1B0B8Y7 A0A1I8P7G4 B4LUL0 A0A194QUC1 I3QC10 A0A194Q1W6 A0A0L0C6A4 A0A1A9XQ71 A0A084VDU8 A0A194PVH3 T1PFM2 A0A194PWK6 A0A182KX90 A0A1I8JTB7 Q5TVS6 A0A1L8DYQ8 A0A0Q9WAE5 A0A194QQB7 A0A182U574 A0A0J9R159 B4HWR7 A0A182QBZ9 A0A182UUQ4 A0A194PXA6 A0A034VW56 Q9VKT9 A0A1Y9J0B7 A0A0A1WMJ9 A0A1L8DPQ4 A0A1L8DPW0 A0A3B0JHG8 A0A1L8DPY1 M9PCU5 B3N5D9 A0A1L8DPM5 Q29KR9 W5J6X8 A0A0J9TKN6 B4P0Q9 M9MRM3 B4GSW4 A0A182J854 A0A3B0KDQ2 A0A0Q5WBH0 A0A194QNQ2 B4MWR9 A0A1Y9H2S0 A0A0R3NZX2 T1EA03 A0A2H1VT83 B3MJK2 A0A0R1DRH5 A0A1L8DQ28 B4MWQ5 A0A0K8W418 A0A1L8DPS2 A0A0Q9WQJ1 A0A182R887 A0A1W4UT02 D2A0X3 B3MJL9 B0W6G8 Q16MC6 A0A0M5J114 B4KKS2 A0A067RCF7 B4GSX7 A0A2J7QW32 A0A1I8MIJ0 A0A194QNR0 A0A1W4VT50 Q9VKS5 Q29KR3 B4Q9R3 B4HWT2 B3N558 B4JE79 A0A3B0JN49 A0A0P8XGJ3 B4P0S2 B4JE91 A0A0K8VTT0
A0A1A9WZU7 A0A1A9UTK2 H9JQH1 A0A2A4K5R5 W8C916 A0A1L8DQ21 A0A1L8EIM3 A0A1L8EII7 A0A1L8EIH0 A0A1L8EID0 A0A2A4K5V7 A0A1B0B8Y7 A0A1I8P7G4 B4LUL0 A0A194QUC1 I3QC10 A0A194Q1W6 A0A0L0C6A4 A0A1A9XQ71 A0A084VDU8 A0A194PVH3 T1PFM2 A0A194PWK6 A0A182KX90 A0A1I8JTB7 Q5TVS6 A0A1L8DYQ8 A0A0Q9WAE5 A0A194QQB7 A0A182U574 A0A0J9R159 B4HWR7 A0A182QBZ9 A0A182UUQ4 A0A194PXA6 A0A034VW56 Q9VKT9 A0A1Y9J0B7 A0A0A1WMJ9 A0A1L8DPQ4 A0A1L8DPW0 A0A3B0JHG8 A0A1L8DPY1 M9PCU5 B3N5D9 A0A1L8DPM5 Q29KR9 W5J6X8 A0A0J9TKN6 B4P0Q9 M9MRM3 B4GSW4 A0A182J854 A0A3B0KDQ2 A0A0Q5WBH0 A0A194QNQ2 B4MWR9 A0A1Y9H2S0 A0A0R3NZX2 T1EA03 A0A2H1VT83 B3MJK2 A0A0R1DRH5 A0A1L8DQ28 B4MWQ5 A0A0K8W418 A0A1L8DPS2 A0A0Q9WQJ1 A0A182R887 A0A1W4UT02 D2A0X3 B3MJL9 B0W6G8 Q16MC6 A0A0M5J114 B4KKS2 A0A067RCF7 B4GSX7 A0A2J7QW32 A0A1I8MIJ0 A0A194QNR0 A0A1W4VT50 Q9VKS5 Q29KR3 B4Q9R3 B4HWT2 B3N558 B4JE79 A0A3B0JN49 A0A0P8XGJ3 B4P0S2 B4JE91 A0A0K8VTT0
Pubmed
22118469
26354079
19121390
24495485
17994087
26108605
+ More
24438588 25315136 20966253 12364791 14747013 17210077 22936249 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 15632085 20920257 23761445 17550304 18362917 19820115 17510324 24845553
24438588 25315136 20966253 12364791 14747013 17210077 22936249 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 15632085 20920257 23761445 17550304 18362917 19820115 17510324 24845553
EMBL
NWSH01005801
PCG63937.1
ODYU01012460
SOQ58781.1
AGBW02009374
OWR50972.1
+ More
KQ460855 KPJ11879.1 KQ459601 KPI94007.1 BABH01004735 BABH01004736 BABH01004737 BABH01011980 NWSH01000114 PCG79409.1 GAMC01007031 JAB99524.1 GFDF01005639 JAV08445.1 GFDG01000330 JAV18469.1 GFDG01000334 JAV18465.1 GFDG01000331 JAV18468.1 GFDG01000332 JAV18467.1 PCG79406.1 JXJN01010106 CH940649 EDW64196.1 KQ461190 KPJ07151.1 JF999959 AFI64313.1 KQ459590 KPI97395.1 JRES01000842 KNC27786.1 ATLV01011799 KE524723 KFB36142.1 KPI97391.1 KA647607 AFP62236.1 KPI97393.1 APCN01002227 AAAB01008820 EAL41497.4 GFDF01002510 JAV11574.1 KRF81490.1 KPJ07150.1 CM002910 KMY89574.1 CH480818 EDW52462.1 AXCN02000090 KPI97394.1 GAKP01013224 JAC45728.1 AE014134 AY089586 AAF52971.1 AAL90324.1 GBXI01014562 JAC99729.1 GFDF01005642 JAV08442.1 GFDF01005643 JAV08441.1 OUUW01000006 SPP81787.1 GFDF01005647 JAV08437.1 AGB92891.1 CH954177 EDV59018.1 GFDF01005646 JAV08438.1 CH379061 EAL33105.2 ADMH02002130 ETN58534.1 KMY89575.1 CM000157 EDW87954.1 ADV37035.1 CH479189 EDW25473.1 SPP81788.1 KQS70596.1 KPJ07153.1 CH963857 EDW76558.1 KRT04741.1 GAMD01001453 JAB00138.1 ODYU01004000 SOQ43434.1 CH902620 EDV32370.2 KRJ97507.1 GFDF01005644 JAV08440.1 EDW76544.2 GDHF01006507 JAI45807.1 GFDF01005645 JAV08439.1 KRF98484.1 KQ971338 EFA01616.1 EDV32387.1 DS231848 EDS36642.1 CH477866 EAT35491.1 CP012523 ALC38094.1 CH933807 EDW12736.1 KK852794 KDR16428.1 EDW25486.1 NEVH01009768 PNF32784.1 KPJ07152.1 BT003518 AAF52985.2 AAO39522.1 EAL33110.2 CM000361 EDX04580.1 KMY89593.1 EDW52477.1 EDV59003.1 CH916368 EDW03599.1 SPP81772.1 KPU73921.1 EDW87967.1 EDW03611.1 GDHF01010028 GDHF01008682 JAI42286.1 JAI43632.1
KQ460855 KPJ11879.1 KQ459601 KPI94007.1 BABH01004735 BABH01004736 BABH01004737 BABH01011980 NWSH01000114 PCG79409.1 GAMC01007031 JAB99524.1 GFDF01005639 JAV08445.1 GFDG01000330 JAV18469.1 GFDG01000334 JAV18465.1 GFDG01000331 JAV18468.1 GFDG01000332 JAV18467.1 PCG79406.1 JXJN01010106 CH940649 EDW64196.1 KQ461190 KPJ07151.1 JF999959 AFI64313.1 KQ459590 KPI97395.1 JRES01000842 KNC27786.1 ATLV01011799 KE524723 KFB36142.1 KPI97391.1 KA647607 AFP62236.1 KPI97393.1 APCN01002227 AAAB01008820 EAL41497.4 GFDF01002510 JAV11574.1 KRF81490.1 KPJ07150.1 CM002910 KMY89574.1 CH480818 EDW52462.1 AXCN02000090 KPI97394.1 GAKP01013224 JAC45728.1 AE014134 AY089586 AAF52971.1 AAL90324.1 GBXI01014562 JAC99729.1 GFDF01005642 JAV08442.1 GFDF01005643 JAV08441.1 OUUW01000006 SPP81787.1 GFDF01005647 JAV08437.1 AGB92891.1 CH954177 EDV59018.1 GFDF01005646 JAV08438.1 CH379061 EAL33105.2 ADMH02002130 ETN58534.1 KMY89575.1 CM000157 EDW87954.1 ADV37035.1 CH479189 EDW25473.1 SPP81788.1 KQS70596.1 KPJ07153.1 CH963857 EDW76558.1 KRT04741.1 GAMD01001453 JAB00138.1 ODYU01004000 SOQ43434.1 CH902620 EDV32370.2 KRJ97507.1 GFDF01005644 JAV08440.1 EDW76544.2 GDHF01006507 JAI45807.1 GFDF01005645 JAV08439.1 KRF98484.1 KQ971338 EFA01616.1 EDV32387.1 DS231848 EDS36642.1 CH477866 EAT35491.1 CP012523 ALC38094.1 CH933807 EDW12736.1 KK852794 KDR16428.1 EDW25486.1 NEVH01009768 PNF32784.1 KPJ07152.1 BT003518 AAF52985.2 AAO39522.1 EAL33110.2 CM000361 EDX04580.1 KMY89593.1 EDW52477.1 EDV59003.1 CH916368 EDW03599.1 SPP81772.1 KPU73921.1 EDW87967.1 EDW03611.1 GDHF01010028 GDHF01008682 JAI42286.1 JAI43632.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000005204
UP000091820
+ More
UP000078200 UP000092460 UP000095300 UP000008792 UP000037069 UP000092443 UP000030765 UP000095301 UP000075882 UP000075840 UP000007062 UP000075902 UP000001292 UP000075886 UP000075903 UP000000803 UP000076407 UP000268350 UP000008711 UP000001819 UP000000673 UP000002282 UP000008744 UP000075880 UP000007798 UP000075884 UP000007801 UP000075900 UP000192221 UP000007266 UP000002320 UP000008820 UP000092553 UP000009192 UP000027135 UP000235965 UP000000304 UP000001070
UP000078200 UP000092460 UP000095300 UP000008792 UP000037069 UP000092443 UP000030765 UP000095301 UP000075882 UP000075840 UP000007062 UP000075902 UP000001292 UP000075886 UP000075903 UP000000803 UP000076407 UP000268350 UP000008711 UP000001819 UP000000673 UP000002282 UP000008744 UP000075880 UP000007798 UP000075884 UP000007801 UP000075900 UP000192221 UP000007266 UP000002320 UP000008820 UP000092553 UP000009192 UP000027135 UP000235965 UP000000304 UP000001070
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2A4IVE2
A0A2H1X0E5
A0A212FB51
A0A194R2Y3
A0A194PL00
H9J9F6
+ More
A0A1A9WZU7 A0A1A9UTK2 H9JQH1 A0A2A4K5R5 W8C916 A0A1L8DQ21 A0A1L8EIM3 A0A1L8EII7 A0A1L8EIH0 A0A1L8EID0 A0A2A4K5V7 A0A1B0B8Y7 A0A1I8P7G4 B4LUL0 A0A194QUC1 I3QC10 A0A194Q1W6 A0A0L0C6A4 A0A1A9XQ71 A0A084VDU8 A0A194PVH3 T1PFM2 A0A194PWK6 A0A182KX90 A0A1I8JTB7 Q5TVS6 A0A1L8DYQ8 A0A0Q9WAE5 A0A194QQB7 A0A182U574 A0A0J9R159 B4HWR7 A0A182QBZ9 A0A182UUQ4 A0A194PXA6 A0A034VW56 Q9VKT9 A0A1Y9J0B7 A0A0A1WMJ9 A0A1L8DPQ4 A0A1L8DPW0 A0A3B0JHG8 A0A1L8DPY1 M9PCU5 B3N5D9 A0A1L8DPM5 Q29KR9 W5J6X8 A0A0J9TKN6 B4P0Q9 M9MRM3 B4GSW4 A0A182J854 A0A3B0KDQ2 A0A0Q5WBH0 A0A194QNQ2 B4MWR9 A0A1Y9H2S0 A0A0R3NZX2 T1EA03 A0A2H1VT83 B3MJK2 A0A0R1DRH5 A0A1L8DQ28 B4MWQ5 A0A0K8W418 A0A1L8DPS2 A0A0Q9WQJ1 A0A182R887 A0A1W4UT02 D2A0X3 B3MJL9 B0W6G8 Q16MC6 A0A0M5J114 B4KKS2 A0A067RCF7 B4GSX7 A0A2J7QW32 A0A1I8MIJ0 A0A194QNR0 A0A1W4VT50 Q9VKS5 Q29KR3 B4Q9R3 B4HWT2 B3N558 B4JE79 A0A3B0JN49 A0A0P8XGJ3 B4P0S2 B4JE91 A0A0K8VTT0
A0A1A9WZU7 A0A1A9UTK2 H9JQH1 A0A2A4K5R5 W8C916 A0A1L8DQ21 A0A1L8EIM3 A0A1L8EII7 A0A1L8EIH0 A0A1L8EID0 A0A2A4K5V7 A0A1B0B8Y7 A0A1I8P7G4 B4LUL0 A0A194QUC1 I3QC10 A0A194Q1W6 A0A0L0C6A4 A0A1A9XQ71 A0A084VDU8 A0A194PVH3 T1PFM2 A0A194PWK6 A0A182KX90 A0A1I8JTB7 Q5TVS6 A0A1L8DYQ8 A0A0Q9WAE5 A0A194QQB7 A0A182U574 A0A0J9R159 B4HWR7 A0A182QBZ9 A0A182UUQ4 A0A194PXA6 A0A034VW56 Q9VKT9 A0A1Y9J0B7 A0A0A1WMJ9 A0A1L8DPQ4 A0A1L8DPW0 A0A3B0JHG8 A0A1L8DPY1 M9PCU5 B3N5D9 A0A1L8DPM5 Q29KR9 W5J6X8 A0A0J9TKN6 B4P0Q9 M9MRM3 B4GSW4 A0A182J854 A0A3B0KDQ2 A0A0Q5WBH0 A0A194QNQ2 B4MWR9 A0A1Y9H2S0 A0A0R3NZX2 T1EA03 A0A2H1VT83 B3MJK2 A0A0R1DRH5 A0A1L8DQ28 B4MWQ5 A0A0K8W418 A0A1L8DPS2 A0A0Q9WQJ1 A0A182R887 A0A1W4UT02 D2A0X3 B3MJL9 B0W6G8 Q16MC6 A0A0M5J114 B4KKS2 A0A067RCF7 B4GSX7 A0A2J7QW32 A0A1I8MIJ0 A0A194QNR0 A0A1W4VT50 Q9VKS5 Q29KR3 B4Q9R3 B4HWT2 B3N558 B4JE79 A0A3B0JN49 A0A0P8XGJ3 B4P0S2 B4JE91 A0A0K8VTT0
PDB
1K8Q
E-value=4.43208e-71,
Score=681
Ontologies
PATHWAY
GO
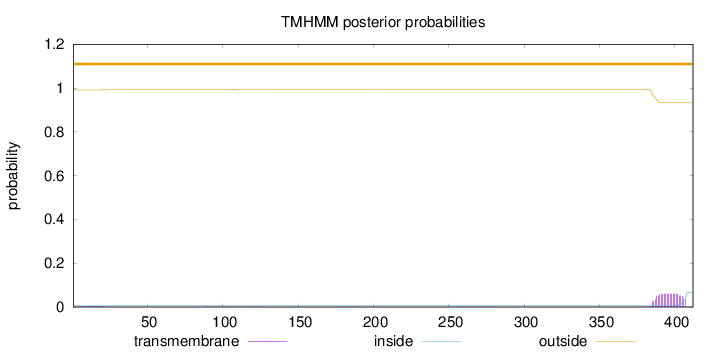
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.957985
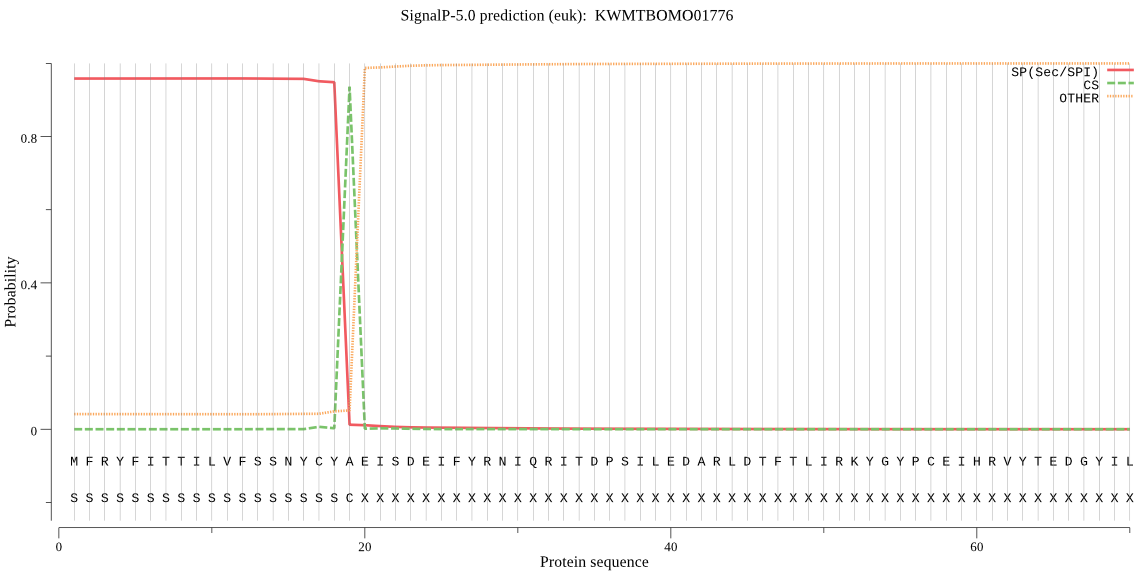
Length:
412
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.30558
Exp number, first 60 AAs:
0.05913
Total prob of N-in:
0.00826
outside
1 - 412
Population Genetic Test Statistics
Pi
232.951927
Theta
170.134661
Tajima's D
1.628932
CLR
0.254702
CSRT
0.814859257037148
Interpretation
Uncertain
