Gene
KWMTBOMO01774
Pre Gene Modal
BGIBMGA014526
Annotation
PREDICTED:_neither_inactivation_nor_afterpotential_protein_C_[Amyelois_transitella]
Full name
Protein Wnt
+ More
Neither inactivation nor afterpotential protein C
Neither inactivation nor afterpotential protein C
Location in the cell
Nuclear Reliability : 3.372
Sequence
CDS
ATGAGATTAGGCTCGTTCCTAATTTACGAAGCCCCTGCACTTATGCGAGTTCGGAACGTGGTTGCCCACTTGTTCTTGTGGAAGGAGTTCAGGCTAGACGTGTTAGTCTGCTGTGGTCTGCTGCAACCAGATATTCTGCAAAGGTATCTTCATGCGCCATCCAAAATGAAAGACGGTCTCAACTTATCGTCCTTGCAAAATCCTTTGGAACGTTACAAACTAGACACTAAACTAGGTTCTGGCATATTTGGGGTAGTTTACAAGGCTACCGATACACAGGCGGCTGGAAAATCCGTTGCTGTTAAAATACAAAAATATACCGATAAAAATGAGGCACACATTCAAAAGGAATACAAAATACTTCGAGATTTCACTACTCATCTTAATCTTATCGACTTTTACGGAGTGTTCTGCGAGAGAGTCGATAATATAAACAAAATATGGTTTGTTCTTGAACTGTGCGAAAACGGATCGGTCATCGATATTGTCAGAAAGCTGAATACGGCAGAAAGAAAAATGTCCGAAGAACACATAGCTTACATCTTAAAATATACAATAAAGGCCCTCGTTTATCTGCACGAGAACAAAGTGATGCATCGGAACATAAGGTGTAGCAATATTTTAATCACAAAAGACGGTGAAGTTAAACTGTCTGATTTTGGACTGTCGTGTAAATTAAGCGGAACACTTGATAAGAGTAAAACGAATGTTGGTTCTCCGAGTTGGATGGCGCCCGAAGTGGTGACTGGCGGAGACGACGGTTACGGCAACAGGGTCGACGTTTGGGCCTTGGGCATAACTACTATAGAGATGGCAGACGGAAAAGCCCCTTACCAGGACATGCATCCGACACGCGCATTGTTTCAAATAGTAAGATGCCCTCCGCCGACCATTTCGAAGCCTTCAATGTGGTCGAACGATATCAACGATTTCATTTCTGAGTGTCTCGAAAAAAATCCGGAACATCGACCTTTCATAATGGAACTGGAAGAGCATCCGTTTATTCTGTCCGTTCCTGAGAACGATTATCACCTCTCAACAGAATTGAAGATACTCGCGAAAGAACTAAGTGGCAAGGTATCGCCAGATAAAACACCAGAACGGATAATAAGAGATGGCTTATTGACAACTGAAGGGAACCCTGAAGCCGAAACGATGCAAGTTGAGGATCTTGCCGCTCTAGAAATATTGACTGAGGAAAACTTACTCAATGAGTTGCAAATAAAATTGATGAAAGGTTCATTTATGTCTTTCGTCGGTGACATTCTACTTATTCTCAACCCCAACACCGATGATGATATTTATAACGAAAACCATCACAAGAAGTATGAGTGCAAATCAAGATCAGATAATGAGCCGCACATATTCGCGGTAGCCGATGGCGCTTACCAAGACGCACTTCATCATAACGAACCACAGCACATCGTGTTCTCTGGAGAAAGCAAATCCGGCAAAACTACGAACATGTTACACGCTCTGTCACATTTGACTCATTTAGGAGCGATGAAGAATAACACAGCAGATAGAATTCGTAAAGCAACTAGTATTATCCAAGCAGCTATTTCTGCCGCTACACCCATAAATGCACATTGCACCAGGGGTATTTTTCAAGTTCAAGTGACATACGGAAGCTCGGGAAAACTGAGCGGCGCTATCTTTTGGTTGTATCAATTAGAAAAGTGGCGAATCTCATCAACAGACATGACCCACGCTAACTTCGACTTAATTTATTATTTCTACGATGCGATGGAAGCGGGAAATCGATTAGATGAACTCTGTTTAGACAAAAACAGAAAACATAGATACTTACGGATATCAGATGAACCTGAAAAAGAATTAAAAGGCGTTAGAGAAAGTGTCGAAAATAACGTTCAAAATTATAAAAATATCATTGAGAACCTCACCGCAGTCGATTGGGAAGAAGAGGACATAGGATTTTTTGAAACAGTTTTAGCGTCGATTTTAGTTCTCGGTAATGTCAGATTCAAAGAAGGAAGGAATGGTTCAACCGAAATTGAAAATCCGGAGGAGGCTAAGAGGGTAGCGAAGCTATTGTCACTGGATGAAACAAAATTCCTTTGGGCTTTATTAAATTACTGTCTTATCGAAAGCGGGACTGCAGTAAAAAGAAAACATACTACAGATGAAGCGAGAGATGCTAGGGACACATTAGCAGGCACTTTATATAAAAGACTCATTGACTGGATGATAAATTTAATTAATTCGAAACTCTCCTTTATGAGATCTGTCTTTGGTGATAAATATTCAGTCAGCTTATTGGATATGTTCGGTTTTGAATGCTATCACAGAAATCGACTAGAGCAATTGATAGTAAATACAACAAATGAACAAATACAGTTCCTTTATAACCAAAGAATTTTCGCTTGGGAAATGCAAGAAGCCACAGAAGAAGGTATCGCAATGCCAGAACTAAGATTTTATGACAACAAAAATTCTGTGGATCAACTCATGGGCAGACCTTTAGGCTTGTTTTATGTATTGGACGAAGCGAGTAGAACTAGTAGTGGACAGGAATTCATCATGAGTACAATAAGGACAACGTGTAAAGGACCGTACATCAAATTATCCGGCAGTCATGAATTCAGTGTAGCTCACTATACAGGCAAAGTGAGCTACGATGCAAGAGAAATGGCAGAAAAGAACAAAGACTTCCTTCCTCCTGAAATGATAGAAACAATGAGGGCATCAGTCAATATAACAGTGCAACAGTTATTTAGAAATAAATTAACGAAAACAGGCAATCTTACGGTTTCGCCAAGTCAGCCTAAAGTCACAGTTGCTAAATCAAAATCAGAAAAGGAATTGGAAAACCTAAAAACTAGGAAATTCAACACGGTCTCCAAAGGGCAATTTTCTCAAGTTCATAGAATGAGAACAGCGGCTGCAACATACAGAGCAACCAGTTTAGAATTATTGAAACAGCTCGCCGTTGGGCCCGGCAGTGGCGGAACGCATTACGTCAGATGCATCAGACCAGATTTGAATGACAATCCCCGAGGTTTCCAAAAAGAGGTCGTGAGACAACAATTAAGAGCCTTGGCTGTTTTAGACACGGCCAAAGCCAGGCAGAGTGGCTTCTCCAGTCGCATACCTTTTGCTGAATTCATAAGGAGGTACCGCTTCTTAGCCTTCGATTTCGACGAAAACGTGGAAATGACAAAAGACAACTGTAGACTACTTATGATAAGGTTAAAAATGGAAGGTTGGGAGTTGGGAAAAACGAAAGTTTTCCTGAAATACTATAATGAAGAATTCTTGGCAAGATTGTACGAAACAAAAGTGAAAAAAATTATAAAAGTGCAATGTATGATGAGAGCTTTCTTGGCCAGAAGAAGGGCGATCAAGAGTAAATCAAAATTATTGCATATCAAAGAACTGAAGAAACAACAATCTCTGAACGTAACTGAGGATGAAGCGGCACTGTTGATTCAAAAAGCATACAGAGGTTATATCGTTCGTAAAGCATACGGACCGCTCATAAACAAATCTACGGGGCAAATTGACGAGGAAACAGCTTCGTTCCTAAAAAGATTCGCAATGAAATGGAAGAGCCGCTCCATATTCCAAGTTCTGCTGCAGTACAGAGCGGTTCGCTATCAGGACTTAGTGCACTTCTCGCAACAAGTGCATATTTACAATCAAGCCGTTTTAGAAGCACTGATGACAACTAGCACATCCGTTTTGCTAGACCGCGTGGACCCGCAAGCTAAAGAAGCCAATTTCCTTGGAACTAGCCCTCCGACCGTCTGGAAGCTACCGTTCAGAATTGATCAACTACAATTCTATGATACGATGCACATGTGCGATCCATCTGCAAAAAGCACCGACACTTTTGCTGGTCAGTACGATTCCGATCAAGAGCAATGGGACGAACCTCTGAAACGACGTTTTAGCGCCACACAGATAGACAGCATCGTAACGGCCACAACTCAAACGCTCATCACCGTGCCGTTCTGCCGAGATCCCACCCAGCCTGTCCCTAATCTGCCTCCAGAAGAAACTATCCAACAGCCACCATCCTCTCCTCGTATCGAGGATGGGTCCCGCAACCGTCCGACGGTCTACAAAAAGAAACACGGTAATGCACCACAAAGTTACTACCAGCCCCCGGAGCCGTGGAACGGTGCCCGCGACGACGACGACGTTCTAGAAAACAGCGCACCCAAATTTAAAAAGACGCACAGCCGAAGTCTTTATTGTATCGACGCTCCAGACATGCAGAATCCGATAAAAGAACTCCAAGCCATGGCGAGATCTACGAATGATTTGACCGAGGACGACGCACCATTCAACTTTCAAGCTATGTTGAAGAGGACGCCAAAAAATAGAGCGTCGATGAAACGACTGGGCGAGATCGACAACGGAACATTGGAGAAGCCGCAGTCTACGCCGAAACGTGCGCCGACACCAAAAGCAAAAGCTCCTCCCCCTCCCGTTCAAGTAACACCATCGAAAGCGCCAAACAAGGTCTTACCACCAACTCCACCACCCAGCGAAGTCACGCCCTCGCCGACACCAAATTCAATGCATAAAGAATTCACCAGGCATGATTCCAAAGATCTCATAATACAAGAATTACAGAAACGAGAATCTAGGCCAGAGTTCATTTACCCGAGCAACAAAGTTGAAAGCGAAAAAATCGAATTAGCCCCCGGTATCACGATTCAGGGGACCGTTACAGATTTATAA
Protein
MRLGSFLIYEAPALMRVRNVVAHLFLWKEFRLDVLVCCGLLQPDILQRYLHAPSKMKDGLNLSSLQNPLERYKLDTKLGSGIFGVVYKATDTQAAGKSVAVKIQKYTDKNEAHIQKEYKILRDFTTHLNLIDFYGVFCERVDNINKIWFVLELCENGSVIDIVRKLNTAERKMSEEHIAYILKYTIKALVYLHENKVMHRNIRCSNILITKDGEVKLSDFGLSCKLSGTLDKSKTNVGSPSWMAPEVVTGGDDGYGNRVDVWALGITTIEMADGKAPYQDMHPTRALFQIVRCPPPTISKPSMWSNDINDFISECLEKNPEHRPFIMELEEHPFILSVPENDYHLSTELKILAKELSGKVSPDKTPERIIRDGLLTTEGNPEAETMQVEDLAALEILTEENLLNELQIKLMKGSFMSFVGDILLILNPNTDDDIYNENHHKKYECKSRSDNEPHIFAVADGAYQDALHHNEPQHIVFSGESKSGKTTNMLHALSHLTHLGAMKNNTADRIRKATSIIQAAISAATPINAHCTRGIFQVQVTYGSSGKLSGAIFWLYQLEKWRISSTDMTHANFDLIYYFYDAMEAGNRLDELCLDKNRKHRYLRISDEPEKELKGVRESVENNVQNYKNIIENLTAVDWEEEDIGFFETVLASILVLGNVRFKEGRNGSTEIENPEEAKRVAKLLSLDETKFLWALLNYCLIESGTAVKRKHTTDEARDARDTLAGTLYKRLIDWMINLINSKLSFMRSVFGDKYSVSLLDMFGFECYHRNRLEQLIVNTTNEQIQFLYNQRIFAWEMQEATEEGIAMPELRFYDNKNSVDQLMGRPLGLFYVLDEASRTSSGQEFIMSTIRTTCKGPYIKLSGSHEFSVAHYTGKVSYDAREMAEKNKDFLPPEMIETMRASVNITVQQLFRNKLTKTGNLTVSPSQPKVTVAKSKSEKELENLKTRKFNTVSKGQFSQVHRMRTAAATYRATSLELLKQLAVGPGSGGTHYVRCIRPDLNDNPRGFQKEVVRQQLRALAVLDTAKARQSGFSSRIPFAEFIRRYRFLAFDFDENVEMTKDNCRLLMIRLKMEGWELGKTKVFLKYYNEEFLARLYETKVKKIIKVQCMMRAFLARRRAIKSKSKLLHIKELKKQQSLNVTEDEAALLIQKAYRGYIVRKAYGPLINKSTGQIDEETASFLKRFAMKWKSRSIFQVLLQYRAVRYQDLVHFSQQVHIYNQAVLEALMTTSTSVLLDRVDPQAKEANFLGTSPPTVWKLPFRIDQLQFYDTMHMCDPSAKSTDTFAGQYDSDQEQWDEPLKRRFSATQIDSIVTATTQTLITVPFCRDPTQPVPNLPPEETIQQPPSSPRIEDGSRNRPTVYKKKHGNAPQSYYQPPEPWNGARDDDDVLENSAPKFKKTHSRSLYCIDAPDMQNPIKELQAMARSTNDLTEDDAPFNFQAMLKRTPKNRASMKRLGEIDNGTLEKPQSTPKRAPTPKAKAPPPPVQVTPSKAPNKVLPPTPPPSEVTPSPTPNSMHKEFTRHDSKDLIIQELQKRESRPEFIYPSNKVESEKIELAPGITIQGTVTDL
Summary
Description
Ligand for members of the frizzled family of seven transmembrane receptors.
Required for photoreceptor cell function. The ninaC proteins combines putative serine/threonine-protein kinase and myosin activities (PubMed:2449973). Essential for the expression and stability of the rtp protein in the photoreceptors (PubMed:20107052). The rtp/ninaC complex is required for stability of inad and inac and the normal termination of phototransduction in the retina (PubMed:20739554).
Required for photoreceptor cell function. The ninaC proteins combines putative serine/threonine-protein kinase and myosin activities (PubMed:2449973). Essential for the expression and stability of the rtp protein in the photoreceptors (PubMed:20107052). The rtp/ninaC complex is required for stability of inad and inac and the normal termination of phototransduction in the retina (PubMed:20739554).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Interacts with rtp.
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the Wnt family.
In the C-terminal section; belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
In the N-terminal section; belongs to the protein kinase superfamily. Ser/Thr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the Wnt family.
In the C-terminal section; belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
In the N-terminal section; belongs to the protein kinase superfamily. Ser/Thr protein kinase family.
Keywords
Actin-binding
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Cytoskeleton
Kinase
Membrane
Motor protein
Myosin
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
Sensory transduction
Transferase
Vision
Feature
chain Neither inactivation nor afterpotential protein C
splice variant In isoform A.
splice variant In isoform A.
Uniprot
H9JYA6
A0A0L7LL24
A0A194PKQ0
A0A212FB55
A0A2A4J3R9
A0A2H1WDP1
+ More
A0A026W0S7 A0A3L8DYX9 A0A195DA37 K7JA59 F4WZ45 W5JRM6 A0A182R1L4 A0A182WGH2 A0A182F0Y9 B0WGM7 Q17NQ0 E9IYU1 A0A1Q3FS76 A0A151XEP3 A0A182NBP7 A0A182GB27 A0A0L7RB11 A0A195CLC5 A0A1Q3FSQ0 A0A182IJF2 A0A158NDP2 A0A182XN71 A0A182HRZ0 Q7Q1L3 A0A182TI09 A0A084WU58 A0A182UZ09 A0A182QQM6 A0A195BQU1 A0A195EYW1 A0A182K2Z8 A0A0M9A245 E2B612 A0A182P5Q9 A0A088A2N1 A0A182KU51 A0A1Q3FS78 A0A182Y0M2 A0A1B6L4F4 A0A154PIA9 A0A1W4X848 A0A1W4WXA5 A0A1B6LPF4 A0A1J1I1S5 A0A2A3E6B9 A0A067RAF4 A0A0A9Z778 A0A1B6CTP3 T1HKH8 E2A0L0 A0A1B6C2U7 A0A1W4VWB1 B4KKC5 B4MZW1 B4LS40 Q29N85 B3N6B9 B4HY52 P10676 A0A0J9QXL9 A0A0L0BS66 B4NZL6 B4JQN5 W8BGP4 A0A3B0K2B3 D6WK70 A0A1A9WUE0 J9JKK8 A0A1I8NV56 A0A1I8N044 A0A336JYY3 A0A0K8UTQ8 A0A1B0BRP9 A0A2P8ZDP2 A0A1A9XYR9 A0A2H8TIA1 A0A182MND6 A0A0K8VRY2 A0A0A1XS93 W8C697 A0A1I8N040 T1PFP8
A0A026W0S7 A0A3L8DYX9 A0A195DA37 K7JA59 F4WZ45 W5JRM6 A0A182R1L4 A0A182WGH2 A0A182F0Y9 B0WGM7 Q17NQ0 E9IYU1 A0A1Q3FS76 A0A151XEP3 A0A182NBP7 A0A182GB27 A0A0L7RB11 A0A195CLC5 A0A1Q3FSQ0 A0A182IJF2 A0A158NDP2 A0A182XN71 A0A182HRZ0 Q7Q1L3 A0A182TI09 A0A084WU58 A0A182UZ09 A0A182QQM6 A0A195BQU1 A0A195EYW1 A0A182K2Z8 A0A0M9A245 E2B612 A0A182P5Q9 A0A088A2N1 A0A182KU51 A0A1Q3FS78 A0A182Y0M2 A0A1B6L4F4 A0A154PIA9 A0A1W4X848 A0A1W4WXA5 A0A1B6LPF4 A0A1J1I1S5 A0A2A3E6B9 A0A067RAF4 A0A0A9Z778 A0A1B6CTP3 T1HKH8 E2A0L0 A0A1B6C2U7 A0A1W4VWB1 B4KKC5 B4MZW1 B4LS40 Q29N85 B3N6B9 B4HY52 P10676 A0A0J9QXL9 A0A0L0BS66 B4NZL6 B4JQN5 W8BGP4 A0A3B0K2B3 D6WK70 A0A1A9WUE0 J9JKK8 A0A1I8NV56 A0A1I8N044 A0A336JYY3 A0A0K8UTQ8 A0A1B0BRP9 A0A2P8ZDP2 A0A1A9XYR9 A0A2H8TIA1 A0A182MND6 A0A0K8VRY2 A0A0A1XS93 W8C697 A0A1I8N040 T1PFP8
EC Number
2.7.11.1
Pubmed
19121390
26227816
26354079
22118469
24508170
30249741
+ More
20075255 21719571 20920257 23761445 17510324 21282665 26483478 21347285 12364791 14747013 17210077 24438588 20798317 20966253 25244985 24845553 25401762 17994087 15632085 18057021 2449973 10731132 12537572 12537569 18327897 20107052 20739554 25822849 22936249 26108605 17550304 24495485 18362917 19820115 25315136 29403074 25830018
20075255 21719571 20920257 23761445 17510324 21282665 26483478 21347285 12364791 14747013 17210077 24438588 20798317 20966253 25244985 24845553 25401762 17994087 15632085 18057021 2449973 10731132 12537572 12537569 18327897 20107052 20739554 25822849 22936249 26108605 17550304 24495485 18362917 19820115 25315136 29403074 25830018
EMBL
BABH01042828
BABH01042829
BABH01042830
JTDY01000778
KOB75911.1
KQ459601
+ More
KPI94011.1 AGBW02009374 OWR50974.1 NWSH01003424 PCG66328.1 ODYU01007965 SOQ51183.1 KK107578 EZA48669.1 QOIP01000002 RLU25647.1 KQ981082 KYN09731.1 AAZX01001625 GL888465 EGI60548.1 ADMH02000623 ETN65570.1 DS231927 EDS27056.1 CH477197 EAT48353.1 GL767051 EFZ14256.1 GFDL01004601 JAV30444.1 KQ982254 KYQ58801.1 JXUM01052119 KQ561720 KXJ77723.1 KQ414617 KOC68107.1 KQ977600 KYN01503.1 GFDL01004602 JAV30443.1 ADTU01012732 ADTU01012733 ADTU01012734 ADTU01012735 ADTU01012736 APCN01001777 AAAB01008980 EAA14178.4 ATLV01027002 KE525421 KFB53752.1 AXCN02000684 KQ976424 KYM88329.1 KQ981906 KYN33316.1 KQ435793 KOX74259.1 GL445900 EFN88873.1 GFDL01004618 JAV30427.1 GEBQ01021466 JAT18511.1 KQ434899 KZC10930.1 GEBQ01014480 JAT25497.1 CVRI01000037 CRK93540.1 KZ288367 PBC26822.1 KK852595 KDR20620.1 GBHO01005985 GBHO01005984 GBHO01005981 JAG37619.1 JAG37620.1 JAG37623.1 GEDC01020389 JAS16909.1 ACPB03023451 GL435626 EFN72913.1 GEDC01029452 JAS07846.1 CH933807 EDW12656.1 CH963920 EDW77896.1 CH940649 EDW64726.1 CH379060 EAL33457.1 CH954177 EDV59206.1 KQS70689.1 CH480818 EDW51982.1 J03131 M20230 M20231 AE014134 AY119496 CM002910 KMY88751.1 JRES01001454 KNC22848.1 CM000157 EDW87765.1 KRJ97381.1 CH916372 EDV99215.1 GAMC01008643 JAB97912.1 OUUW01000006 SPP82020.1 KQ971342 EFA03943.2 ABLF02030790 ABLF02030792 UFQS01000004 UFQT01000004 SSW96792.1 SSX17179.1 GDHF01022338 JAI29976.1 JXJN01019182 JXJN01019183 JXJN01019184 JXJN01019185 JXJN01019186 JXJN01019187 PYGN01000087 PSN54627.1 GFXV01002068 MBW13873.1 AXCM01001833 GDHF01010677 JAI41637.1 GBXI01000884 JAD13408.1 GAMC01008641 JAB97914.1 KA647489 AFP62118.1
KPI94011.1 AGBW02009374 OWR50974.1 NWSH01003424 PCG66328.1 ODYU01007965 SOQ51183.1 KK107578 EZA48669.1 QOIP01000002 RLU25647.1 KQ981082 KYN09731.1 AAZX01001625 GL888465 EGI60548.1 ADMH02000623 ETN65570.1 DS231927 EDS27056.1 CH477197 EAT48353.1 GL767051 EFZ14256.1 GFDL01004601 JAV30444.1 KQ982254 KYQ58801.1 JXUM01052119 KQ561720 KXJ77723.1 KQ414617 KOC68107.1 KQ977600 KYN01503.1 GFDL01004602 JAV30443.1 ADTU01012732 ADTU01012733 ADTU01012734 ADTU01012735 ADTU01012736 APCN01001777 AAAB01008980 EAA14178.4 ATLV01027002 KE525421 KFB53752.1 AXCN02000684 KQ976424 KYM88329.1 KQ981906 KYN33316.1 KQ435793 KOX74259.1 GL445900 EFN88873.1 GFDL01004618 JAV30427.1 GEBQ01021466 JAT18511.1 KQ434899 KZC10930.1 GEBQ01014480 JAT25497.1 CVRI01000037 CRK93540.1 KZ288367 PBC26822.1 KK852595 KDR20620.1 GBHO01005985 GBHO01005984 GBHO01005981 JAG37619.1 JAG37620.1 JAG37623.1 GEDC01020389 JAS16909.1 ACPB03023451 GL435626 EFN72913.1 GEDC01029452 JAS07846.1 CH933807 EDW12656.1 CH963920 EDW77896.1 CH940649 EDW64726.1 CH379060 EAL33457.1 CH954177 EDV59206.1 KQS70689.1 CH480818 EDW51982.1 J03131 M20230 M20231 AE014134 AY119496 CM002910 KMY88751.1 JRES01001454 KNC22848.1 CM000157 EDW87765.1 KRJ97381.1 CH916372 EDV99215.1 GAMC01008643 JAB97912.1 OUUW01000006 SPP82020.1 KQ971342 EFA03943.2 ABLF02030790 ABLF02030792 UFQS01000004 UFQT01000004 SSW96792.1 SSX17179.1 GDHF01022338 JAI29976.1 JXJN01019182 JXJN01019183 JXJN01019184 JXJN01019185 JXJN01019186 JXJN01019187 PYGN01000087 PSN54627.1 GFXV01002068 MBW13873.1 AXCM01001833 GDHF01010677 JAI41637.1 GBXI01000884 JAD13408.1 GAMC01008641 JAB97914.1 KA647489 AFP62118.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000007151
UP000218220
UP000053097
+ More
UP000279307 UP000078492 UP000002358 UP000007755 UP000000673 UP000075900 UP000075920 UP000069272 UP000002320 UP000008820 UP000075809 UP000075884 UP000069940 UP000249989 UP000053825 UP000078542 UP000075880 UP000005205 UP000076407 UP000075840 UP000007062 UP000075902 UP000030765 UP000075903 UP000075886 UP000078540 UP000078541 UP000075881 UP000053105 UP000008237 UP000075885 UP000005203 UP000075882 UP000076408 UP000076502 UP000192223 UP000183832 UP000242457 UP000027135 UP000015103 UP000000311 UP000192221 UP000009192 UP000007798 UP000008792 UP000001819 UP000008711 UP000001292 UP000000803 UP000037069 UP000002282 UP000001070 UP000268350 UP000007266 UP000091820 UP000007819 UP000095300 UP000095301 UP000092460 UP000245037 UP000092443 UP000075883
UP000279307 UP000078492 UP000002358 UP000007755 UP000000673 UP000075900 UP000075920 UP000069272 UP000002320 UP000008820 UP000075809 UP000075884 UP000069940 UP000249989 UP000053825 UP000078542 UP000075880 UP000005205 UP000076407 UP000075840 UP000007062 UP000075902 UP000030765 UP000075903 UP000075886 UP000078540 UP000078541 UP000075881 UP000053105 UP000008237 UP000075885 UP000005203 UP000075882 UP000076408 UP000076502 UP000192223 UP000183832 UP000242457 UP000027135 UP000015103 UP000000311 UP000192221 UP000009192 UP000007798 UP000008792 UP000001819 UP000008711 UP000001292 UP000000803 UP000037069 UP000002282 UP000001070 UP000268350 UP000007266 UP000091820 UP000007819 UP000095300 UP000095301 UP000092460 UP000245037 UP000092443 UP000075883
Pfam
Interpro
IPR000719
Prot_kinase_dom
+ More
IPR000048 IQ_motif_EF-hand-BS
IPR011009 Kinase-like_dom_sf
IPR027417 P-loop_NTPase
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001609 Myosin_head_motor_dom
IPR017441 Protein_kinase_ATP_BS
IPR036961 Kinesin_motor_dom_sf
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR005817 Wnt
IPR018161 Wnt_CS
IPR008271 Ser/Thr_kinase_AS
IPR024276 CCAP
IPR000048 IQ_motif_EF-hand-BS
IPR011009 Kinase-like_dom_sf
IPR027417 P-loop_NTPase
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001609 Myosin_head_motor_dom
IPR017441 Protein_kinase_ATP_BS
IPR036961 Kinesin_motor_dom_sf
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR005817 Wnt
IPR018161 Wnt_CS
IPR008271 Ser/Thr_kinase_AS
IPR024276 CCAP
Gene 3D
ProteinModelPortal
H9JYA6
A0A0L7LL24
A0A194PKQ0
A0A212FB55
A0A2A4J3R9
A0A2H1WDP1
+ More
A0A026W0S7 A0A3L8DYX9 A0A195DA37 K7JA59 F4WZ45 W5JRM6 A0A182R1L4 A0A182WGH2 A0A182F0Y9 B0WGM7 Q17NQ0 E9IYU1 A0A1Q3FS76 A0A151XEP3 A0A182NBP7 A0A182GB27 A0A0L7RB11 A0A195CLC5 A0A1Q3FSQ0 A0A182IJF2 A0A158NDP2 A0A182XN71 A0A182HRZ0 Q7Q1L3 A0A182TI09 A0A084WU58 A0A182UZ09 A0A182QQM6 A0A195BQU1 A0A195EYW1 A0A182K2Z8 A0A0M9A245 E2B612 A0A182P5Q9 A0A088A2N1 A0A182KU51 A0A1Q3FS78 A0A182Y0M2 A0A1B6L4F4 A0A154PIA9 A0A1W4X848 A0A1W4WXA5 A0A1B6LPF4 A0A1J1I1S5 A0A2A3E6B9 A0A067RAF4 A0A0A9Z778 A0A1B6CTP3 T1HKH8 E2A0L0 A0A1B6C2U7 A0A1W4VWB1 B4KKC5 B4MZW1 B4LS40 Q29N85 B3N6B9 B4HY52 P10676 A0A0J9QXL9 A0A0L0BS66 B4NZL6 B4JQN5 W8BGP4 A0A3B0K2B3 D6WK70 A0A1A9WUE0 J9JKK8 A0A1I8NV56 A0A1I8N044 A0A336JYY3 A0A0K8UTQ8 A0A1B0BRP9 A0A2P8ZDP2 A0A1A9XYR9 A0A2H8TIA1 A0A182MND6 A0A0K8VRY2 A0A0A1XS93 W8C697 A0A1I8N040 T1PFP8
A0A026W0S7 A0A3L8DYX9 A0A195DA37 K7JA59 F4WZ45 W5JRM6 A0A182R1L4 A0A182WGH2 A0A182F0Y9 B0WGM7 Q17NQ0 E9IYU1 A0A1Q3FS76 A0A151XEP3 A0A182NBP7 A0A182GB27 A0A0L7RB11 A0A195CLC5 A0A1Q3FSQ0 A0A182IJF2 A0A158NDP2 A0A182XN71 A0A182HRZ0 Q7Q1L3 A0A182TI09 A0A084WU58 A0A182UZ09 A0A182QQM6 A0A195BQU1 A0A195EYW1 A0A182K2Z8 A0A0M9A245 E2B612 A0A182P5Q9 A0A088A2N1 A0A182KU51 A0A1Q3FS78 A0A182Y0M2 A0A1B6L4F4 A0A154PIA9 A0A1W4X848 A0A1W4WXA5 A0A1B6LPF4 A0A1J1I1S5 A0A2A3E6B9 A0A067RAF4 A0A0A9Z778 A0A1B6CTP3 T1HKH8 E2A0L0 A0A1B6C2U7 A0A1W4VWB1 B4KKC5 B4MZW1 B4LS40 Q29N85 B3N6B9 B4HY52 P10676 A0A0J9QXL9 A0A0L0BS66 B4NZL6 B4JQN5 W8BGP4 A0A3B0K2B3 D6WK70 A0A1A9WUE0 J9JKK8 A0A1I8NV56 A0A1I8N044 A0A336JYY3 A0A0K8UTQ8 A0A1B0BRP9 A0A2P8ZDP2 A0A1A9XYR9 A0A2H8TIA1 A0A182MND6 A0A0K8VRY2 A0A0A1XS93 W8C697 A0A1I8N040 T1PFP8
Ontologies
GO
GO:0003774
GO:0003779
GO:0004672
GO:0016459
GO:0005524
GO:0004713
GO:0005102
GO:0005576
GO:0007275
GO:0016055
GO:0033227
GO:0016062
GO:0007010
GO:0045494
GO:0006886
GO:0035091
GO:0004674
GO:0016059
GO:0007604
GO:0005634
GO:1990146
GO:0005737
GO:0033583
GO:0005516
GO:0016027
GO:0016461
GO:0042385
GO:0007601
GO:0016028
GO:0007602
GO:0007603
GO:0006468
GO:0042623
GO:0009416
GO:0008104
GO:0005515
GO:0006810
GO:0006397
GO:0004651
Topology
Subcellular location
Cytoplasm Localizes in the cytoplasm or punctate structures within the nuclei in the absence of rtp. Co-localizes with rtp in the rhabdomere membrane. With evidence from 10 publications.
Cytoskeleton Localizes in the cytoplasm or punctate structures within the nuclei in the absence of rtp. Co-localizes with rtp in the rhabdomere membrane. With evidence from 10 publications.
Nucleus Localizes in the cytoplasm or punctate structures within the nuclei in the absence of rtp. Co-localizes with rtp in the rhabdomere membrane. With evidence from 10 publications.
Membrane Localizes in the cytoplasm or punctate structures within the nuclei in the absence of rtp. Co-localizes with rtp in the rhabdomere membrane. With evidence from 10 publications.
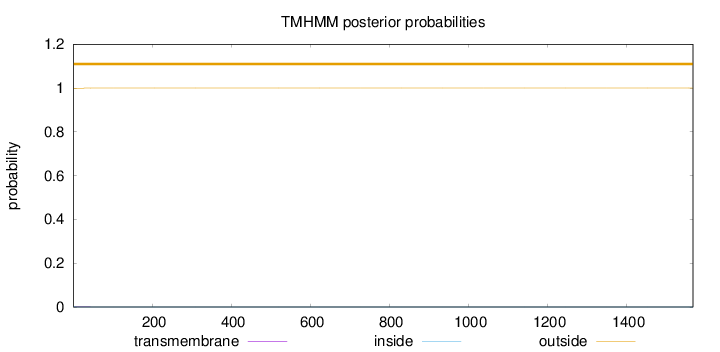
Length:
1569
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02595
Exp number, first 60 AAs:
0.02547
Total prob of N-in:
0.00118
outside
1 - 1569
Population Genetic Test Statistics
Pi
240.845887
Theta
203.325204
Tajima's D
-1.465269
CLR
1.870102
CSRT
0.0624968751562422
Interpretation
Uncertain
