Gene
KWMTBOMO01773
Annotation
PREDICTED:_protein_Wnt-10-like?_partial_[Bombyx_mori]
Full name
Protein Wnt
+ More
Protein Wnt-10a
Protein Wnt-10a
Location in the cell
Extracellular Reliability : 2.207 PlasmaMembrane Reliability : 1.662
Sequence
CDS
ATGGAGGTGCGGTGCAAATGTCATGGTTTGTCCGGAAGCTGTCAACTGCGGACATGTTGGAGGACCACGCCTGATTTCAGAGTGGTCGCTTCAACTATCAAAAGACAGTATCGGAAGGCATTAGTTGCAGCGCAAGAGGGACAGATTAATAACGGCATGTCAGTATTGAAGGGTAGACCGAGGGGTAGGAGGAAGAGCAAACCACGACCGGCGCCAAAGACAAGTCTGTTGTTTTTTGAACGGTCACCTAATTTCTGTGAAGTGGACCCCAGGACGGACTCGGCCGGCACAAGTGGGAGAGTGTGTCGTGTGGGACGACCCTCGAGGTCTGGCGCTTGCGAGCTCCTGTGCTGCGGTCGGGGTCACTCTCTCGTCAGGAAATCCAGCATTAAGCCCTGCAACTGTACCTTTCACTGGTGCTGCACTGTCGACTGTCAACGGTGCAGAGACGATAGATGGATCGCTGAAATTGAACAACTCAAAAAGGTGCTGTAA
Protein
MEVRCKCHGLSGSCQLRTCWRTTPDFRVVASTIKRQYRKALVAAQEGQINNGMSVLKGRPRGRRKSKPRPAPKTSLLFFERSPNFCEVDPRTDSAGTSGRVCRVGRPSRSGACELLCCGRGHSLVRKSSIKPCNCTFHWCCTVDCQRCRDDRWIAEIEQLKKVL
Summary
Description
Ligand for members of the frizzled family of seven transmembrane receptors.
Ligand for members of the frizzled family of seven transmembrane receptors (Probable). Required for normal tooth development (PubMed:29178643). Regulates the expression of genes involved in tooth development (PubMed:29178643).
Ligand for members of the frizzled family of seven transmembrane receptors (Probable). Required for normal tooth development (PubMed:29178643). Regulates the expression of genes involved in tooth development (PubMed:29178643).
Miscellaneous
Wnt10a overexpression in embryos results in loss of anterior neural identity, causing a phenotype with small or no eyes.
Similarity
Belongs to the Wnt family.
Keywords
Complete proteome
Developmental protein
Disulfide bond
Extracellular matrix
Glycoprotein
Lipoprotein
Reference proteome
Secreted
Signal
Wnt signaling pathway
Feature
chain Protein Wnt
Uniprot
A0A2H1WNJ4
A0A2A4J4F7
A0A194PL05
A0A194R258
A0A212FB54
A0A077DH97
+ More
A0A2R7WJD0 H3BB92 P79856 Q9PVT4 A0A1S3NWD7 A0A060VS93 A0A1V4K7P4 G1KMN3 T1HKG5 A0A3P8YFJ0 A0A3B3SK21 A0A3P8WW94 W5MDP0 A0A091I7F7 F6SDS7 A0A3B3Y5W7 W5MDL7 Q3L249 A0A1A6G307 A0A3P9PMZ5 A0A3B5M4Z1 G1NFT1 A0A3P9PN69 H2S5F3 A0A3B4FB28 A0A091FKL6 H3DI33 A0A091VYP1 A0A091E6A1 A0A087QTS5 A0A093G2U2 A0A2J7RGB2 A0A091N7G1 A0A1S3PLK3 A0A093R9J2 A0A099Z7K4 A0A091PPU6 U3JNI5 A0A2I0LIK5 A0A346FPZ3 A0A0Q3PT81 A0A1V4K642 A0A3B4EGQ0 A0A1L2A1N5 A0A2I0UKI2 A0A226NNY1 A0A3L8SN20 A0A3M0L0V5 A0A226PBI7 A0A2P4T077 Q60GF6 A0A3N0Z251 A0A3B5BNJ7 A0A3P8P6E3 A0A3P9AYH6 G1NFT5 A0A3B4YE50 A0A0P7URS5 A4ZGN7 H3C786 A0A1U7Q659 B7ZQU5 A0A218VAZ4 A0A0S7IX60 H3BIB0 I3J9F4 A0A1W5ASP3 B0V1G8 A0A3B3R208 A0A3B4YL29 H3DC73 Q4RW24 A0A3B4UB81 A0A3P9PMY1 A0A3B3UX37 G3T9L8 A0A0F8CJL4 A0A1W4ZDN4 A0A3B5QKE5 A0A2Y9DKT0 A0A3P8SPC5 A0A3B1JZM6 A0A3P9KLN9 A0A3P9IZU5 H2TUW6 A0A3B3X597 A0A096M425 A0A3B4D0Z2 P43446 Q9CUV1 A0A087XFF5 A0A0P7X879 H2MVK9
A0A2R7WJD0 H3BB92 P79856 Q9PVT4 A0A1S3NWD7 A0A060VS93 A0A1V4K7P4 G1KMN3 T1HKG5 A0A3P8YFJ0 A0A3B3SK21 A0A3P8WW94 W5MDP0 A0A091I7F7 F6SDS7 A0A3B3Y5W7 W5MDL7 Q3L249 A0A1A6G307 A0A3P9PMZ5 A0A3B5M4Z1 G1NFT1 A0A3P9PN69 H2S5F3 A0A3B4FB28 A0A091FKL6 H3DI33 A0A091VYP1 A0A091E6A1 A0A087QTS5 A0A093G2U2 A0A2J7RGB2 A0A091N7G1 A0A1S3PLK3 A0A093R9J2 A0A099Z7K4 A0A091PPU6 U3JNI5 A0A2I0LIK5 A0A346FPZ3 A0A0Q3PT81 A0A1V4K642 A0A3B4EGQ0 A0A1L2A1N5 A0A2I0UKI2 A0A226NNY1 A0A3L8SN20 A0A3M0L0V5 A0A226PBI7 A0A2P4T077 Q60GF6 A0A3N0Z251 A0A3B5BNJ7 A0A3P8P6E3 A0A3P9AYH6 G1NFT5 A0A3B4YE50 A0A0P7URS5 A4ZGN7 H3C786 A0A1U7Q659 B7ZQU5 A0A218VAZ4 A0A0S7IX60 H3BIB0 I3J9F4 A0A1W5ASP3 B0V1G8 A0A3B3R208 A0A3B4YL29 H3DC73 Q4RW24 A0A3B4UB81 A0A3P9PMY1 A0A3B3UX37 G3T9L8 A0A0F8CJL4 A0A1W4ZDN4 A0A3B5QKE5 A0A2Y9DKT0 A0A3P8SPC5 A0A3B1JZM6 A0A3P9KLN9 A0A3P9IZU5 H2TUW6 A0A3B3X597 A0A096M425 A0A3B4D0Z2 P43446 Q9CUV1 A0A087XFF5 A0A0P7X879 H2MVK9
Pubmed
26354079
22118469
25196151
9215903
9022051
24755649
+ More
25069045 29240929 24487278 20431018 16258938 20838655 21551351 15496914 23371554 29567137 30282656 24621616 15592404 15789446 25186727 17436276 27762356 23594743 25835551 23542700 25329095 17554307 8330668 29178643 10349636 11042159 11076861 11217851 12466851 16141073
25069045 29240929 24487278 20431018 16258938 20838655 21551351 15496914 23371554 29567137 30282656 24621616 15592404 15789446 25186727 17436276 27762356 23594743 25835551 23542700 25329095 17554307 8330668 29178643 10349636 11042159 11076861 11217851 12466851 16141073
EMBL
ODYU01009908
SOQ54641.1
NWSH01003424
PCG66330.1
KQ459601
KPI94012.1
+ More
KQ460855 KPJ11873.1 AGBW02009374 OWR50975.1 KJ906609 AIL33838.1 KK854927 PTY19736.1 AFYH01036103 AFYH01036104 AFYH01036105 AFYH01036106 AFYH01036107 AFYH01036108 AFYH01036109 AFYH01036110 AFYH01036111 U65428 AAB38531.1 AB026729 BAA85879.1 FR904292 CDQ57828.1 LSYS01004331 OPJ79917.1 ACPB03023451 AHAT01028894 AHAT01028895 KL218243 KFP03315.1 AAMC01028450 AAMC01028451 AY753295 AAW81997.1 LZPO01107543 OBS60581.1 KL447081 KFO69694.1 KL410197 KFQ94713.1 KK717809 KFO52766.1 KL225902 KFM04629.1 KL215153 KFV63403.1 NEVH01004401 PNF39863.1 KK842622 KFP84789.1 KL225119 KFW67659.1 KL890375 KGL77746.1 KK677284 KFQ09942.1 AGTO01001151 AKCR02000392 PKK17203.1 MF416116 AXN72354.1 LMAW01001113 KQK84443.1 OPJ79916.1 KU169900 ALO81631.1 KZ505705 PKU46552.1 MCFN01000003 OXB69201.1 QUSF01000012 RLW04917.1 QRBI01000096 RMC18641.1 AWGT02000105 OXB77114.1 PPHD01014126 POI29761.1 AADN05000350 AB177400 BAD61009.1 RJVU01016332 ROL52352.1 JARO02002572 KPP72365.1 DQ658155 ABG49498.1 BC169932 BC169934 CM004482 AAI69932.1 OCT63407.1 MUZQ01000015 OWK63215.1 GBYX01423809 JAO57422.1 AFYH01001812 AFYH01001813 AERX01009451 CR847976 CAAE01014991 CAG07408.1 KQ041905 KKF21274.1 AYCK01009421 U02544 AK013836 BAB29012.1 JARO02003374 KPP70623.1
KQ460855 KPJ11873.1 AGBW02009374 OWR50975.1 KJ906609 AIL33838.1 KK854927 PTY19736.1 AFYH01036103 AFYH01036104 AFYH01036105 AFYH01036106 AFYH01036107 AFYH01036108 AFYH01036109 AFYH01036110 AFYH01036111 U65428 AAB38531.1 AB026729 BAA85879.1 FR904292 CDQ57828.1 LSYS01004331 OPJ79917.1 ACPB03023451 AHAT01028894 AHAT01028895 KL218243 KFP03315.1 AAMC01028450 AAMC01028451 AY753295 AAW81997.1 LZPO01107543 OBS60581.1 KL447081 KFO69694.1 KL410197 KFQ94713.1 KK717809 KFO52766.1 KL225902 KFM04629.1 KL215153 KFV63403.1 NEVH01004401 PNF39863.1 KK842622 KFP84789.1 KL225119 KFW67659.1 KL890375 KGL77746.1 KK677284 KFQ09942.1 AGTO01001151 AKCR02000392 PKK17203.1 MF416116 AXN72354.1 LMAW01001113 KQK84443.1 OPJ79916.1 KU169900 ALO81631.1 KZ505705 PKU46552.1 MCFN01000003 OXB69201.1 QUSF01000012 RLW04917.1 QRBI01000096 RMC18641.1 AWGT02000105 OXB77114.1 PPHD01014126 POI29761.1 AADN05000350 AB177400 BAD61009.1 RJVU01016332 ROL52352.1 JARO02002572 KPP72365.1 DQ658155 ABG49498.1 BC169932 BC169934 CM004482 AAI69932.1 OCT63407.1 MUZQ01000015 OWK63215.1 GBYX01423809 JAO57422.1 AFYH01001812 AFYH01001813 AERX01009451 CR847976 CAAE01014991 CAG07408.1 KQ041905 KKF21274.1 AYCK01009421 U02544 AK013836 BAB29012.1 JARO02003374 KPP70623.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000008672
UP000087266
+ More
UP000193380 UP000190648 UP000001646 UP000015103 UP000265140 UP000261540 UP000265120 UP000018468 UP000054308 UP000008143 UP000261480 UP000092124 UP000242638 UP000261380 UP000001645 UP000005226 UP000261460 UP000053760 UP000007303 UP000053283 UP000052976 UP000053286 UP000053875 UP000235965 UP000054081 UP000053641 UP000016665 UP000053872 UP000051836 UP000261440 UP000198323 UP000276834 UP000269221 UP000198419 UP000000539 UP000261400 UP000265100 UP000265160 UP000261360 UP000034805 UP000189706 UP000186698 UP000197619 UP000005207 UP000192224 UP000000437 UP000261420 UP000261500 UP000007646 UP000002852 UP000248480 UP000265080 UP000018467 UP000265180 UP000265200 UP000028760 UP000001038
UP000193380 UP000190648 UP000001646 UP000015103 UP000265140 UP000261540 UP000265120 UP000018468 UP000054308 UP000008143 UP000261480 UP000092124 UP000242638 UP000261380 UP000001645 UP000005226 UP000261460 UP000053760 UP000007303 UP000053283 UP000052976 UP000053286 UP000053875 UP000235965 UP000054081 UP000053641 UP000016665 UP000053872 UP000051836 UP000261440 UP000198323 UP000276834 UP000269221 UP000198419 UP000000539 UP000261400 UP000265100 UP000265160 UP000261360 UP000034805 UP000189706 UP000186698 UP000197619 UP000005207 UP000192224 UP000000437 UP000261420 UP000261500 UP000007646 UP000002852 UP000248480 UP000265080 UP000018467 UP000265180 UP000265200 UP000028760 UP000001038
PRIDE
Pfam
PF00110 wnt
ProteinModelPortal
A0A2H1WNJ4
A0A2A4J4F7
A0A194PL05
A0A194R258
A0A212FB54
A0A077DH97
+ More
A0A2R7WJD0 H3BB92 P79856 Q9PVT4 A0A1S3NWD7 A0A060VS93 A0A1V4K7P4 G1KMN3 T1HKG5 A0A3P8YFJ0 A0A3B3SK21 A0A3P8WW94 W5MDP0 A0A091I7F7 F6SDS7 A0A3B3Y5W7 W5MDL7 Q3L249 A0A1A6G307 A0A3P9PMZ5 A0A3B5M4Z1 G1NFT1 A0A3P9PN69 H2S5F3 A0A3B4FB28 A0A091FKL6 H3DI33 A0A091VYP1 A0A091E6A1 A0A087QTS5 A0A093G2U2 A0A2J7RGB2 A0A091N7G1 A0A1S3PLK3 A0A093R9J2 A0A099Z7K4 A0A091PPU6 U3JNI5 A0A2I0LIK5 A0A346FPZ3 A0A0Q3PT81 A0A1V4K642 A0A3B4EGQ0 A0A1L2A1N5 A0A2I0UKI2 A0A226NNY1 A0A3L8SN20 A0A3M0L0V5 A0A226PBI7 A0A2P4T077 Q60GF6 A0A3N0Z251 A0A3B5BNJ7 A0A3P8P6E3 A0A3P9AYH6 G1NFT5 A0A3B4YE50 A0A0P7URS5 A4ZGN7 H3C786 A0A1U7Q659 B7ZQU5 A0A218VAZ4 A0A0S7IX60 H3BIB0 I3J9F4 A0A1W5ASP3 B0V1G8 A0A3B3R208 A0A3B4YL29 H3DC73 Q4RW24 A0A3B4UB81 A0A3P9PMY1 A0A3B3UX37 G3T9L8 A0A0F8CJL4 A0A1W4ZDN4 A0A3B5QKE5 A0A2Y9DKT0 A0A3P8SPC5 A0A3B1JZM6 A0A3P9KLN9 A0A3P9IZU5 H2TUW6 A0A3B3X597 A0A096M425 A0A3B4D0Z2 P43446 Q9CUV1 A0A087XFF5 A0A0P7X879 H2MVK9
A0A2R7WJD0 H3BB92 P79856 Q9PVT4 A0A1S3NWD7 A0A060VS93 A0A1V4K7P4 G1KMN3 T1HKG5 A0A3P8YFJ0 A0A3B3SK21 A0A3P8WW94 W5MDP0 A0A091I7F7 F6SDS7 A0A3B3Y5W7 W5MDL7 Q3L249 A0A1A6G307 A0A3P9PMZ5 A0A3B5M4Z1 G1NFT1 A0A3P9PN69 H2S5F3 A0A3B4FB28 A0A091FKL6 H3DI33 A0A091VYP1 A0A091E6A1 A0A087QTS5 A0A093G2U2 A0A2J7RGB2 A0A091N7G1 A0A1S3PLK3 A0A093R9J2 A0A099Z7K4 A0A091PPU6 U3JNI5 A0A2I0LIK5 A0A346FPZ3 A0A0Q3PT81 A0A1V4K642 A0A3B4EGQ0 A0A1L2A1N5 A0A2I0UKI2 A0A226NNY1 A0A3L8SN20 A0A3M0L0V5 A0A226PBI7 A0A2P4T077 Q60GF6 A0A3N0Z251 A0A3B5BNJ7 A0A3P8P6E3 A0A3P9AYH6 G1NFT5 A0A3B4YE50 A0A0P7URS5 A4ZGN7 H3C786 A0A1U7Q659 B7ZQU5 A0A218VAZ4 A0A0S7IX60 H3BIB0 I3J9F4 A0A1W5ASP3 B0V1G8 A0A3B3R208 A0A3B4YL29 H3DC73 Q4RW24 A0A3B4UB81 A0A3P9PMY1 A0A3B3UX37 G3T9L8 A0A0F8CJL4 A0A1W4ZDN4 A0A3B5QKE5 A0A2Y9DKT0 A0A3P8SPC5 A0A3B1JZM6 A0A3P9KLN9 A0A3P9IZU5 H2TUW6 A0A3B3X597 A0A096M425 A0A3B4D0Z2 P43446 Q9CUV1 A0A087XFF5 A0A0P7X879 H2MVK9
PDB
6AHY
E-value=2.85036e-25,
Score=281
Ontologies
PATHWAY
GO
PANTHER
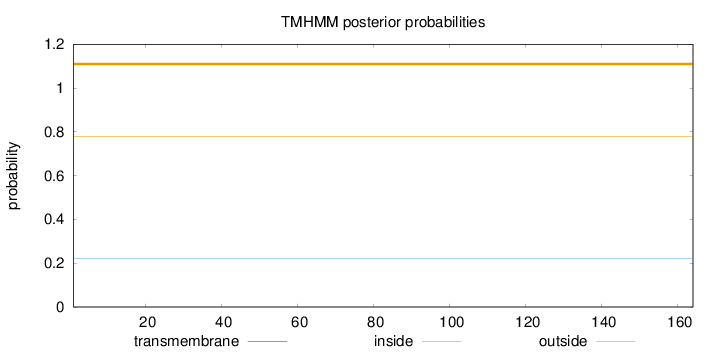
Topology
Subcellular location
Secreted
Extracellular space
Extracellular matrix
Length:
164
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00016
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.22278
outside
1 - 164
Population Genetic Test Statistics
Pi
258.713451
Theta
181.027063
Tajima's D
1.640808
CLR
0.09124
CSRT
0.818209089545523
Interpretation
Uncertain
