Gene
KWMTBOMO01772
Pre Gene Modal
BGIBMGA006004
Annotation
PREDICTED:_protein_Wnt-6_[Bombyx_mori]
Full name
Protein Wnt
Location in the cell
Cytoplasmic Reliability : 1.818 Extracellular Reliability : 1.884
Sequence
CDS
ATGCCCAGCGAAGCTGACGGGGCGTCCGGTAGCGCTGTGATCCTGGATCCTCGCATGGTGTGCAAGAAAAATCGTAGGACACGAGGTAGACTGGCTCAGATCTGCAGAAATGAAACCGGCTTGGTGAAGGAGATCACTAAGGGCGTAACCTTGGGAGCCACCGAGTGCGCCCATCAGTTCAGGAGCCGCCGCTGGAACTGTACTACGCAGAGGCGGAGCATGAGGAAAATATTAATGAGAGATACGAGAGAAACTGGTTTCGTGAACGCCATAACAGCGGCTGGTGTCACCTTCTCCATCACAAGAGCTTGTACTGCTGGGTCTCTTCTGGAATGCTCCTGCGAAAAGGGCATACCAAAACCTCGTCGTGGTCATGATCCGGTGGACCCACCGCCGACACAAGAGTCCAAGAACAAATGGCAATGGGGCGGATGCAGTGACAACGTACGGTTTGGCCTCAAGAAGTCCAGAGAATTCATGGACAGTCGCTACAGGAAACGGAGCGACATTAAAACTCTCATCAAACTACACAACCACAACGCTGGGAGGTTGGCCATTAAGAACAATATGAAGATAGAGTGTAAATGCCACGGACTTTCCGGTTCCTGTACCTTAAGGACTTGCTGGTGGAGAATGCCTACATTCAGGGAAGTAGGAGATAGACTGCGTGACAGATTTGAAGGTGCCGCTAAGGTGATTGCCAGCAACGACGGTGACAGCTTCATGCCAGAGAGCCCGAGCATAAAGAAGCCAGGCAAGAAAGACATTATCTACTCAGAAGAGTCGCCGGACTTTTGTGCGCCAAATTTGAAGACCGGCTCTCTAGGCACCCAGGGAAGACAGTGCAATATCAGCTCTGCCGGCACCGACGGTTGTGATCAAATGTGCTGCCGACGAGGCTACGTGGAAACAACCATTAACGAAACGGAGAACTGCAACTGCCAATTCAAATGGTGCTGCGAAGTTATATGTGAAACATGCAACGTAAAACGAAACATACAAACATGCCTTTAA
Protein
MPSEADGASGSAVILDPRMVCKKNRRTRGRLAQICRNETGLVKEITKGVTLGATECAHQFRSRRWNCTTQRRSMRKILMRDTRETGFVNAITAAGVTFSITRACTAGSLLECSCEKGIPKPRRGHDPVDPPPTQESKNKWQWGGCSDNVRFGLKKSREFMDSRYRKRSDIKTLIKLHNHNAGRLAIKNNMKIECKCHGLSGSCTLRTCWWRMPTFREVGDRLRDRFEGAAKVIASNDGDSFMPESPSIKKPGKKDIIYSEESPDFCAPNLKTGSLGTQGRQCNISSAGTDGCDQMCCRRGYVETTINETENCNCQFKWCCEVICETCNVKRNIQTCL
Summary
Description
Ligand for members of the frizzled family of seven transmembrane receptors.
Similarity
Belongs to the Wnt family.
Feature
chain Protein Wnt
Uniprot
A0A1E1WNB5
A0A2A4JYE0
A0A077DFB1
A0A2H1VV44
A0A212EY02
A0A0L7LHV4
+ More
A0A194R2E6 A0A194PSB3 A0A0C9RP62 A0A151XEV6 A0A195CN42 A0A158NDP3 A0A195D9Y8 A0A1W4X838 A0A195BP99 A0A0L7RB86 A0A195EZK4 A0A3L8DYV3 A0A088A2M8 K7JA61 A0A154PHX6 A0A232EXY3 A0A097ZRW5 A0A0L0BS63 A0A084WU50 A0A1B0FN37 A0A1B0BRQ4 A0A0N0BG43 D6WK67 B0WGM5 A0A1B0CVR8 E5Q9G8 A0A2L2YFF0 A0A2L2YFG7 E9G212 A0A0N7ZMB5 A0A0P6BLC0 E0VF96 A0A1S3NWG0 W5MDK0 F1RAB5 A0A0N8C4K3 F1R205 A0A1S3NWS0 A0A210R2V8 I3J9F3 A0A2D0R1U9 A0A3B3R0N1 A0A3B4FDN7 A0A3P8P6I5 A0A3P9AXV2 T1KGX8 A0A3P9HDP7 H2MVL0 A0A3P9KLV0 H3BC38 A0A3P9HDS3 H9GL01 A0A3B3C2B6 A0A2U9C8T2 A0A3B1JXM0 B0FK92 A0A3B4AF92 B0FK91 A0A2P4SHC5 A0A091LRG7 A0A1A8AFH4 A0A2I4BW50 A0A091E7W5 A0A1L8EVQ7 A0A3B4YKZ2 A0A3B4U9W6 A0A226PAQ0 A0A0Q3UTN4 A0A3P9A4H9 A0A1S3PLQ0 A0A226NPG8 G1NFU4 Q3C2H6 U3JNI4 A0A091FGG0 A0A3L8SMC9 H0Z5I6 A0A3B5BDE8 A0A1W4Z9F3 A0A096M6L0 A0A093NSC8 A0A3B3W0Q6 A0A3B3Y5X2 A0A087XF76 A0A091RKA8 A0A093PUU7 A0A099ZZ44 A0A2I0UKJ2 A0A0S1NF08 A0A3P8SPL5 A0A1V4K6B9 M3ZHV4 A0A3P8WTP5
A0A194R2E6 A0A194PSB3 A0A0C9RP62 A0A151XEV6 A0A195CN42 A0A158NDP3 A0A195D9Y8 A0A1W4X838 A0A195BP99 A0A0L7RB86 A0A195EZK4 A0A3L8DYV3 A0A088A2M8 K7JA61 A0A154PHX6 A0A232EXY3 A0A097ZRW5 A0A0L0BS63 A0A084WU50 A0A1B0FN37 A0A1B0BRQ4 A0A0N0BG43 D6WK67 B0WGM5 A0A1B0CVR8 E5Q9G8 A0A2L2YFF0 A0A2L2YFG7 E9G212 A0A0N7ZMB5 A0A0P6BLC0 E0VF96 A0A1S3NWG0 W5MDK0 F1RAB5 A0A0N8C4K3 F1R205 A0A1S3NWS0 A0A210R2V8 I3J9F3 A0A2D0R1U9 A0A3B3R0N1 A0A3B4FDN7 A0A3P8P6I5 A0A3P9AXV2 T1KGX8 A0A3P9HDP7 H2MVL0 A0A3P9KLV0 H3BC38 A0A3P9HDS3 H9GL01 A0A3B3C2B6 A0A2U9C8T2 A0A3B1JXM0 B0FK92 A0A3B4AF92 B0FK91 A0A2P4SHC5 A0A091LRG7 A0A1A8AFH4 A0A2I4BW50 A0A091E7W5 A0A1L8EVQ7 A0A3B4YKZ2 A0A3B4U9W6 A0A226PAQ0 A0A0Q3UTN4 A0A3P9A4H9 A0A1S3PLQ0 A0A226NPG8 G1NFU4 Q3C2H6 U3JNI4 A0A091FGG0 A0A3L8SMC9 H0Z5I6 A0A3B5BDE8 A0A1W4Z9F3 A0A096M6L0 A0A093NSC8 A0A3B3W0Q6 A0A3B3Y5X2 A0A087XF76 A0A091RKA8 A0A093PUU7 A0A099ZZ44 A0A2I0UKJ2 A0A0S1NF08 A0A3P8SPL5 A0A1V4K6B9 M3ZHV4 A0A3P8WTP5
Pubmed
25196151
22118469
26227816
26354079
21347285
30249741
+ More
20075255 28648823 26108605 24438588 18362917 19820115 21122121 26561354 21292972 20566863 23594743 28812685 25186727 29240929 17554307 9215903 21881562 29451363 25329095 18224714 25463417 27762356 24621616 25069045 20838655 15592404 23499659 30282656 20360741 23542700 24487278
20075255 28648823 26108605 24438588 18362917 19820115 21122121 26561354 21292972 20566863 23594743 28812685 25186727 29240929 17554307 9215903 21881562 29451363 25329095 18224714 25463417 27762356 24621616 25069045 20838655 15592404 23499659 30282656 20360741 23542700 24487278
EMBL
GDQN01002703
JAT88351.1
NWSH01000422
PCG76503.1
KJ906608
AIL33837.1
+ More
ODYU01004605 SOQ44668.1 AGBW02011632 OWR46372.1 JTDY01001064 KOB74985.1 KQ460855 KPJ11872.1 KQ459601 KPI94015.1 GBYB01008946 JAG78713.1 KQ982254 KYQ58808.1 KQ977600 KYN01509.1 ADTU01012741 ADTU01012742 ADTU01012743 ADTU01012744 ADTU01012745 ADTU01012746 KQ981082 KYN09735.1 KQ976424 KYM88335.1 KQ414617 KOC68103.1 KQ981906 KYN33309.1 QOIP01000002 RLU25650.1 AAZX01005056 AAZX01016321 KQ434899 KZC10928.1 NNAY01001700 OXU23183.1 HG529211 CDI40102.1 JRES01001454 KNC22843.1 ATLV01027000 KE525421 KFB53744.1 CCAG010003942 CCAG010003943 CCAG010003944 JXJN01019200 JXJN01019201 JXJN01019202 KQ435793 KOX74261.1 KQ971342 EFA04620.2 DS231927 EDS27054.1 AJWK01031052 AJWK01031053 HQ650545 ADR79164.1 IAAA01026288 IAAA01026291 LAA06874.1 IAAA01026289 LAA06868.1 GL732530 EFX86167.1 GDIP01231417 JAI91984.1 GDIP01020259 JAM83456.1 DS235107 EEB12052.1 AHAT01028894 AHAT01028895 CR847976 CU151893 GDIQ01115435 JAL36291.1 NEDP02000710 OWF55275.1 AERX01009450 CAEY01000072 AFYH01036118 AFYH01036119 AFYH01036120 AFYH01036121 CP026256 AWP12868.1 EU332159 ABY53107.1 EU332158 CM004483 ABY53106.1 OCT61015.1 PPHD01048393 POI23512.1 KK762434 KFP45376.1 HADY01014500 SBP52985.1 KK717809 KFO52767.1 CM004482 OCT63410.1 AWGT02000105 OXB77113.1 LMAW01001113 KQK84442.1 MCFN01000003 OXB69200.1 AADN05000350 AB199733 BAE46751.1 AGTO01001151 KL447081 KFO69695.1 QUSF01000012 RLW04742.1 ABQF01010768 ABQF01010769 ABQF01010770 AYCK01009419 AYCK01009420 KL225119 KFW67658.1 KK820778 KFQ40016.1 KL670757 KFW80151.1 KL869907 KGL86408.1 KZ505705 PKU46553.1 KT818601 ALL53300.1 LSYS01004331 OPJ79915.1
ODYU01004605 SOQ44668.1 AGBW02011632 OWR46372.1 JTDY01001064 KOB74985.1 KQ460855 KPJ11872.1 KQ459601 KPI94015.1 GBYB01008946 JAG78713.1 KQ982254 KYQ58808.1 KQ977600 KYN01509.1 ADTU01012741 ADTU01012742 ADTU01012743 ADTU01012744 ADTU01012745 ADTU01012746 KQ981082 KYN09735.1 KQ976424 KYM88335.1 KQ414617 KOC68103.1 KQ981906 KYN33309.1 QOIP01000002 RLU25650.1 AAZX01005056 AAZX01016321 KQ434899 KZC10928.1 NNAY01001700 OXU23183.1 HG529211 CDI40102.1 JRES01001454 KNC22843.1 ATLV01027000 KE525421 KFB53744.1 CCAG010003942 CCAG010003943 CCAG010003944 JXJN01019200 JXJN01019201 JXJN01019202 KQ435793 KOX74261.1 KQ971342 EFA04620.2 DS231927 EDS27054.1 AJWK01031052 AJWK01031053 HQ650545 ADR79164.1 IAAA01026288 IAAA01026291 LAA06874.1 IAAA01026289 LAA06868.1 GL732530 EFX86167.1 GDIP01231417 JAI91984.1 GDIP01020259 JAM83456.1 DS235107 EEB12052.1 AHAT01028894 AHAT01028895 CR847976 CU151893 GDIQ01115435 JAL36291.1 NEDP02000710 OWF55275.1 AERX01009450 CAEY01000072 AFYH01036118 AFYH01036119 AFYH01036120 AFYH01036121 CP026256 AWP12868.1 EU332159 ABY53107.1 EU332158 CM004483 ABY53106.1 OCT61015.1 PPHD01048393 POI23512.1 KK762434 KFP45376.1 HADY01014500 SBP52985.1 KK717809 KFO52767.1 CM004482 OCT63410.1 AWGT02000105 OXB77113.1 LMAW01001113 KQK84442.1 MCFN01000003 OXB69200.1 AADN05000350 AB199733 BAE46751.1 AGTO01001151 KL447081 KFO69695.1 QUSF01000012 RLW04742.1 ABQF01010768 ABQF01010769 ABQF01010770 AYCK01009419 AYCK01009420 KL225119 KFW67658.1 KK820778 KFQ40016.1 KL670757 KFW80151.1 KL869907 KGL86408.1 KZ505705 PKU46553.1 KT818601 ALL53300.1 LSYS01004331 OPJ79915.1
Proteomes
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
UP000075809
+ More
UP000078542 UP000005205 UP000078492 UP000192223 UP000078540 UP000053825 UP000078541 UP000279307 UP000005203 UP000002358 UP000076502 UP000215335 UP000037069 UP000030765 UP000092444 UP000092460 UP000053105 UP000007266 UP000002320 UP000092461 UP000000305 UP000009046 UP000087266 UP000018468 UP000000437 UP000242188 UP000005207 UP000221080 UP000261540 UP000261460 UP000265100 UP000265160 UP000015104 UP000265200 UP000001038 UP000265180 UP000008672 UP000001646 UP000261560 UP000246464 UP000018467 UP000261520 UP000186698 UP000192220 UP000052976 UP000261360 UP000261420 UP000198419 UP000051836 UP000265140 UP000198323 UP000001645 UP000000539 UP000016665 UP000053760 UP000276834 UP000007754 UP000261400 UP000192224 UP000028760 UP000054081 UP000261500 UP000261480 UP000053258 UP000053858 UP000265080 UP000190648 UP000002852 UP000265120
UP000078542 UP000005205 UP000078492 UP000192223 UP000078540 UP000053825 UP000078541 UP000279307 UP000005203 UP000002358 UP000076502 UP000215335 UP000037069 UP000030765 UP000092444 UP000092460 UP000053105 UP000007266 UP000002320 UP000092461 UP000000305 UP000009046 UP000087266 UP000018468 UP000000437 UP000242188 UP000005207 UP000221080 UP000261540 UP000261460 UP000265100 UP000265160 UP000015104 UP000265200 UP000001038 UP000265180 UP000008672 UP000001646 UP000261560 UP000246464 UP000018467 UP000261520 UP000186698 UP000192220 UP000052976 UP000261360 UP000261420 UP000198419 UP000051836 UP000265140 UP000198323 UP000001645 UP000000539 UP000016665 UP000053760 UP000276834 UP000007754 UP000261400 UP000192224 UP000028760 UP000054081 UP000261500 UP000261480 UP000053258 UP000053858 UP000265080 UP000190648 UP000002852 UP000265120
Pfam
PF00110 wnt
ProteinModelPortal
A0A1E1WNB5
A0A2A4JYE0
A0A077DFB1
A0A2H1VV44
A0A212EY02
A0A0L7LHV4
+ More
A0A194R2E6 A0A194PSB3 A0A0C9RP62 A0A151XEV6 A0A195CN42 A0A158NDP3 A0A195D9Y8 A0A1W4X838 A0A195BP99 A0A0L7RB86 A0A195EZK4 A0A3L8DYV3 A0A088A2M8 K7JA61 A0A154PHX6 A0A232EXY3 A0A097ZRW5 A0A0L0BS63 A0A084WU50 A0A1B0FN37 A0A1B0BRQ4 A0A0N0BG43 D6WK67 B0WGM5 A0A1B0CVR8 E5Q9G8 A0A2L2YFF0 A0A2L2YFG7 E9G212 A0A0N7ZMB5 A0A0P6BLC0 E0VF96 A0A1S3NWG0 W5MDK0 F1RAB5 A0A0N8C4K3 F1R205 A0A1S3NWS0 A0A210R2V8 I3J9F3 A0A2D0R1U9 A0A3B3R0N1 A0A3B4FDN7 A0A3P8P6I5 A0A3P9AXV2 T1KGX8 A0A3P9HDP7 H2MVL0 A0A3P9KLV0 H3BC38 A0A3P9HDS3 H9GL01 A0A3B3C2B6 A0A2U9C8T2 A0A3B1JXM0 B0FK92 A0A3B4AF92 B0FK91 A0A2P4SHC5 A0A091LRG7 A0A1A8AFH4 A0A2I4BW50 A0A091E7W5 A0A1L8EVQ7 A0A3B4YKZ2 A0A3B4U9W6 A0A226PAQ0 A0A0Q3UTN4 A0A3P9A4H9 A0A1S3PLQ0 A0A226NPG8 G1NFU4 Q3C2H6 U3JNI4 A0A091FGG0 A0A3L8SMC9 H0Z5I6 A0A3B5BDE8 A0A1W4Z9F3 A0A096M6L0 A0A093NSC8 A0A3B3W0Q6 A0A3B3Y5X2 A0A087XF76 A0A091RKA8 A0A093PUU7 A0A099ZZ44 A0A2I0UKJ2 A0A0S1NF08 A0A3P8SPL5 A0A1V4K6B9 M3ZHV4 A0A3P8WTP5
A0A194R2E6 A0A194PSB3 A0A0C9RP62 A0A151XEV6 A0A195CN42 A0A158NDP3 A0A195D9Y8 A0A1W4X838 A0A195BP99 A0A0L7RB86 A0A195EZK4 A0A3L8DYV3 A0A088A2M8 K7JA61 A0A154PHX6 A0A232EXY3 A0A097ZRW5 A0A0L0BS63 A0A084WU50 A0A1B0FN37 A0A1B0BRQ4 A0A0N0BG43 D6WK67 B0WGM5 A0A1B0CVR8 E5Q9G8 A0A2L2YFF0 A0A2L2YFG7 E9G212 A0A0N7ZMB5 A0A0P6BLC0 E0VF96 A0A1S3NWG0 W5MDK0 F1RAB5 A0A0N8C4K3 F1R205 A0A1S3NWS0 A0A210R2V8 I3J9F3 A0A2D0R1U9 A0A3B3R0N1 A0A3B4FDN7 A0A3P8P6I5 A0A3P9AXV2 T1KGX8 A0A3P9HDP7 H2MVL0 A0A3P9KLV0 H3BC38 A0A3P9HDS3 H9GL01 A0A3B3C2B6 A0A2U9C8T2 A0A3B1JXM0 B0FK92 A0A3B4AF92 B0FK91 A0A2P4SHC5 A0A091LRG7 A0A1A8AFH4 A0A2I4BW50 A0A091E7W5 A0A1L8EVQ7 A0A3B4YKZ2 A0A3B4U9W6 A0A226PAQ0 A0A0Q3UTN4 A0A3P9A4H9 A0A1S3PLQ0 A0A226NPG8 G1NFU4 Q3C2H6 U3JNI4 A0A091FGG0 A0A3L8SMC9 H0Z5I6 A0A3B5BDE8 A0A1W4Z9F3 A0A096M6L0 A0A093NSC8 A0A3B3W0Q6 A0A3B3Y5X2 A0A087XF76 A0A091RKA8 A0A093PUU7 A0A099ZZ44 A0A2I0UKJ2 A0A0S1NF08 A0A3P8SPL5 A0A1V4K6B9 M3ZHV4 A0A3P8WTP5
PDB
6AHY
E-value=6.5902e-67,
Score=644
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
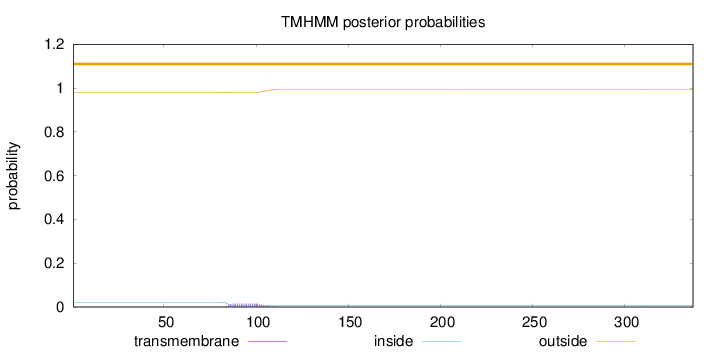
Length:
337
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31747
Exp number, first 60 AAs:
0.00053
Total prob of N-in:
0.02064
outside
1 - 337
Population Genetic Test Statistics
Pi
214.765792
Theta
185.035306
Tajima's D
-1.592793
CLR
0.615308
CSRT
0.047697615119244
Interpretation
Uncertain
