Gene
KWMTBOMO01770
Pre Gene Modal
BGIBMGA006145
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_Wnt-4_[Bombyx_mori]
Full name
Protein Wnt
+ More
Protein Wnt-4
Protein Wnt-4
Alternative Name
dWnt-4
Location in the cell
Mitochondrial Reliability : 1.488
Sequence
CDS
ATGCGTGGTAGTAATGTTCTATCGTTGTTGATTTTGTGTGCCGTTTCTATACCAGCCACGAAAAATGTTAATGGATCGCAACCAATTCTACAAAAGACGATTGAAGCTCTTTTCAGTGATCGCTCGAACATAATCTACCCGGGTTCATGTAAATTGTTTATAGCCTCGAAACGACAAGCGAAAATCTGTACGAGAGAAAAGAAATTTTTCGAGATTTTGACGAACGCGAAGAAACAGGCCATCAGCGCCTGCGAAGAGAGCTTCCAGTACGACAGATGGAACTGTTCACTAATATACAATGGGAAGGCGAAAAAGAGAATTTTTAACAAAATCTATAGAGAGACCGCTTTCATTCACGCCCTGCTGGCGGCATCGTTGACTCATGCGGTCGCGAAAGCCTGCGCTTCCGGAGAGCTGAAGAGCTGCTCATGCGTCGGTAGCTACAGAAAGAACACCACATGGAAAGTCAACGGGTGCGGCGACGACTTCAAGCAAGGCAAGAGAATGATGAAGAACTTTCTGGATTTCAATCACGCCGGCAACGACCAAATCGCTGAAACATTAAAACAAGATGTGATGATTGGTGTTGACTCCATCGGAGAGCAGATGAGGGAGGTGTGTAAATGCCACGGTTTTTCCGGTTCCTGCACAACGAAGACTTGCTGGAAGCGACTGGGGCCGTTCAACTCTGCCATGGGATTGTTAAAGAAGCACTACCATCATGCCGTGAAGAAGAAGCTATTGAATTACACGACCAAAAGAGCTATAACACCGCAAGCTAAGAAGAAAATGAAATCTGACGAGAAATACCTGGTTTATCTGCAGAAGACTCCAAATCTGTGCGTTTGGACCAAAGGTAGAACGTGTAAGAACAGACATAATTGTGCCACGCTGTGTTGCGGGAGGGGCTATTTGACTACCAAGAAGAATGTGACGTCGCGGTGCAAATGCAGGATGGTGAATTGTTGTTTCGTTGAGTGCGATACTTGTGTTAAAGAAGCGGAAATATTCACGTGTAAATAA
Protein
MRGSNVLSLLILCAVSIPATKNVNGSQPILQKTIEALFSDRSNIIYPGSCKLFIASKRQAKICTREKKFFEILTNAKKQAISACEESFQYDRWNCSLIYNGKAKKRIFNKIYRETAFIHALLAASLTHAVAKACASGELKSCSCVGSYRKNTTWKVNGCGDDFKQGKRMMKNFLDFNHAGNDQIAETLKQDVMIGVDSIGEQMREVCKCHGFSGSCTTKTCWKRLGPFNSAMGLLKKHYHHAVKKKLLNYTTKRAITPQAKKKMKSDEKYLVYLQKTPNLCVWTKGRTCKNRHNCATLCCGRGYLTTKKNVTSRCKCRMVNCCFVECDTCVKEAEIFTCK
Summary
Description
Ligand for members of the frizzled family of seven transmembrane receptors.
Binds as a ligand to a family of frizzled seven-transmembrane receptors and acts through a cascade of genes on the nucleus. Acts downstream of homeotic complex genes in the visceral mesoderm and is required for embryonic segmentation. Also required for cell movement and FAK regulation during ovarian morphogenesis.
Binds as a ligand to a family of frizzled seven-transmembrane receptors and acts through a cascade of genes on the nucleus. Acts downstream of homeotic complex genes in the visceral mesoderm and is required for embryonic segmentation. Also required for cell movement and FAK regulation during ovarian morphogenesis.
Similarity
Belongs to the Wnt family.
Keywords
Complete proteome
Developmental protein
Disulfide bond
Extracellular matrix
Glycoprotein
Lipoprotein
Reference proteome
Secreted
Signal
Wnt signaling pathway
Feature
chain Protein Wnt
Uniprot
A0A2H1V5X9
A0A2A4JWX0
A0A212EY09
A0A0L7KV16
A0A182J4L2
A0A182HRY4
+ More
Q17NQ4 A0A182XN77 A0A084WU42 Q16S11 A0A182V617 A0A182GZF1 A0A182QGS0 Q7PM75 A0A182K4F2 A0A182WGH8 A0A182P4Q9 A0A182NBQ3 A0A182TXB1 A0A182R3S5 W5JGY4 B0WGM3 A0A182F0Y2 A0A182YRM0 A0A182MG79 A0A182T9X4 A0A182KU44 A0A1W4VP50 A0A1W4VAJ5 A0A1I8MBQ3 A0A1A9WUF1 A0A1W4VBW8 B4LS45 B4MZW5 A0A3B0KEL3 A0A1A9YDM9 A0A1B0BKX4 A0A0R3NX81 B3N6A9 A0A0M3QTF8 A0A0L0BQQ3 A0A0R3NRN4 B4KKD1 B4Q5E0 B4NZM3 A0A1I8NSI0 A0A0J9QXL2 D2NUH8 P40589 B3MK95 A0A3B0JI52 A0A1A9V1H2 B4G848 B5DHZ8 B4HY45 B4JQN1 A0A0K8U2M1 A0A1A9Z613 W8C1N2 A0A0G3FA69 A0A0A1X1W6 A0A1J1I182 H2Y6S0 W5N397 G1KSZ4 A0A0P7WIA1 H3A9K0 H0WTI3 H3D297 A0A2K5XIG2 Q4S992 I3IYD6 A0A2Y9KWZ8 A0A3B4FPD0 H2Y6R9 H2UHV8 M3YA59 A0A3B4V283 A0A3B3RN63 A0A3P8NMX0 A0A3P9D6W4 F1N7G3 A0A3B3Z8A1 F7APE5 A0A3Q0FYK0 A0A2K5P6W2 A0A2K6LUB5 A0A2K6PMR6 A0A2K6CMG5 A0A2K5TKC3 A0A2I3N5M7 A0A2K5P6S0 A0A2K6CMG2 A0A096P675 A0A3B3RPE7 A0A0D9QUS2 G7PV23 G7NJ57 A0A2K5HBR8
Q17NQ4 A0A182XN77 A0A084WU42 Q16S11 A0A182V617 A0A182GZF1 A0A182QGS0 Q7PM75 A0A182K4F2 A0A182WGH8 A0A182P4Q9 A0A182NBQ3 A0A182TXB1 A0A182R3S5 W5JGY4 B0WGM3 A0A182F0Y2 A0A182YRM0 A0A182MG79 A0A182T9X4 A0A182KU44 A0A1W4VP50 A0A1W4VAJ5 A0A1I8MBQ3 A0A1A9WUF1 A0A1W4VBW8 B4LS45 B4MZW5 A0A3B0KEL3 A0A1A9YDM9 A0A1B0BKX4 A0A0R3NX81 B3N6A9 A0A0M3QTF8 A0A0L0BQQ3 A0A0R3NRN4 B4KKD1 B4Q5E0 B4NZM3 A0A1I8NSI0 A0A0J9QXL2 D2NUH8 P40589 B3MK95 A0A3B0JI52 A0A1A9V1H2 B4G848 B5DHZ8 B4HY45 B4JQN1 A0A0K8U2M1 A0A1A9Z613 W8C1N2 A0A0G3FA69 A0A0A1X1W6 A0A1J1I182 H2Y6S0 W5N397 G1KSZ4 A0A0P7WIA1 H3A9K0 H0WTI3 H3D297 A0A2K5XIG2 Q4S992 I3IYD6 A0A2Y9KWZ8 A0A3B4FPD0 H2Y6R9 H2UHV8 M3YA59 A0A3B4V283 A0A3B3RN63 A0A3P8NMX0 A0A3P9D6W4 F1N7G3 A0A3B3Z8A1 F7APE5 A0A3Q0FYK0 A0A2K5P6W2 A0A2K6LUB5 A0A2K6PMR6 A0A2K6CMG5 A0A2K5TKC3 A0A2I3N5M7 A0A2K5P6S0 A0A2K6CMG2 A0A096P675 A0A3B3RPE7 A0A0D9QUS2 G7PV23 G7NJ57 A0A2K5HBR8
Pubmed
22118469
26227816
17510324
24438588
26483478
12364791
+ More
20920257 23761445 25244985 20966253 25315136 17994087 15632085 26108605 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11970894 7867502 12537569 24495485 26034272 25830018 9215903 15496914 25186727 21551351 29240929 19393038 25463417 19892987 25362486 22002653
20920257 23761445 25244985 20966253 25315136 17994087 15632085 26108605 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11970894 7867502 12537569 24495485 26034272 25830018 9215903 15496914 25186727 21551351 29240929 19393038 25463417 19892987 25362486 22002653
EMBL
ODYU01000848
SOQ36228.1
NWSH01000422
PCG76507.1
AGBW02011632
OWR46375.1
+ More
JTDY01005461 KOB66975.1 APCN01001776 CH477197 EAT48349.1 ATLV01026996 KE525421 KFB53736.1 CH477689 EAT37257.1 JXUM01099512 JXUM01099513 KQ564497 KXJ72185.1 AXCN02000684 AAAB01008980 EAA14176.5 ADMH02001427 ETN62583.1 DS231927 EDS27052.1 AXCM01000768 CH940649 EDW64731.2 CH963920 EDW77900.2 OUUW01000006 SPP82028.1 JXJN01016095 CH379060 KRT03853.1 CH954177 EDV59196.1 CP012523 ALC38809.1 JRES01001508 KNC22390.1 KRT03854.1 CH933807 EDW12662.2 CM000361 EDX04073.1 CM000157 EDW87772.1 CM002910 KMY88746.1 BT120121 AE014134 ADB27992.1 AGB92723.1 AF533773 AY095078 CH902620 EDV32479.1 SPP82027.1 CH479180 EDW28528.1 EDY69935.1 CH480818 EDW51975.1 CH916372 EDV99211.1 GDHF01031598 GDHF01016176 GDHF01009761 JAI20716.1 JAI36138.1 JAI42553.1 GAMC01003477 JAC03079.1 KP966547 AKJ79162.1 GBXI01009170 JAD05122.1 CVRI01000037 CRK93514.1 AHAT01023061 JARO02010201 KPP60877.1 AFYH01094405 AAQR03042343 CAAE01014700 CAG02790.1 AERX01007626 AEYP01079267 AQIA01028175 AQIA01028176 AQIA01028177 AHZZ02010485 AQIB01154041 AQIB01154042 CM001291 EHH58222.1 CM001268 EHH25079.1
JTDY01005461 KOB66975.1 APCN01001776 CH477197 EAT48349.1 ATLV01026996 KE525421 KFB53736.1 CH477689 EAT37257.1 JXUM01099512 JXUM01099513 KQ564497 KXJ72185.1 AXCN02000684 AAAB01008980 EAA14176.5 ADMH02001427 ETN62583.1 DS231927 EDS27052.1 AXCM01000768 CH940649 EDW64731.2 CH963920 EDW77900.2 OUUW01000006 SPP82028.1 JXJN01016095 CH379060 KRT03853.1 CH954177 EDV59196.1 CP012523 ALC38809.1 JRES01001508 KNC22390.1 KRT03854.1 CH933807 EDW12662.2 CM000361 EDX04073.1 CM000157 EDW87772.1 CM002910 KMY88746.1 BT120121 AE014134 ADB27992.1 AGB92723.1 AF533773 AY095078 CH902620 EDV32479.1 SPP82027.1 CH479180 EDW28528.1 EDY69935.1 CH480818 EDW51975.1 CH916372 EDV99211.1 GDHF01031598 GDHF01016176 GDHF01009761 JAI20716.1 JAI36138.1 JAI42553.1 GAMC01003477 JAC03079.1 KP966547 AKJ79162.1 GBXI01009170 JAD05122.1 CVRI01000037 CRK93514.1 AHAT01023061 JARO02010201 KPP60877.1 AFYH01094405 AAQR03042343 CAAE01014700 CAG02790.1 AERX01007626 AEYP01079267 AQIA01028175 AQIA01028176 AQIA01028177 AHZZ02010485 AQIB01154041 AQIB01154042 CM001291 EHH58222.1 CM001268 EHH25079.1
Proteomes
UP000218220
UP000007151
UP000037510
UP000075880
UP000075840
UP000008820
+ More
UP000076407 UP000030765 UP000075903 UP000069940 UP000249989 UP000075886 UP000007062 UP000075881 UP000075920 UP000075885 UP000075884 UP000075902 UP000075900 UP000000673 UP000002320 UP000069272 UP000076408 UP000075883 UP000075901 UP000075882 UP000192221 UP000095301 UP000091820 UP000008792 UP000007798 UP000268350 UP000092443 UP000092460 UP000001819 UP000008711 UP000092553 UP000037069 UP000009192 UP000000304 UP000002282 UP000095300 UP000000803 UP000007801 UP000078200 UP000008744 UP000001292 UP000001070 UP000092445 UP000183832 UP000007875 UP000018468 UP000001646 UP000034805 UP000192224 UP000008672 UP000005225 UP000007303 UP000233140 UP000005207 UP000248482 UP000261460 UP000005226 UP000000715 UP000261420 UP000261540 UP000265100 UP000265160 UP000009136 UP000261520 UP000002281 UP000189705 UP000233060 UP000233180 UP000233200 UP000233120 UP000233100 UP000028761 UP000029965 UP000009130 UP000233080
UP000076407 UP000030765 UP000075903 UP000069940 UP000249989 UP000075886 UP000007062 UP000075881 UP000075920 UP000075885 UP000075884 UP000075902 UP000075900 UP000000673 UP000002320 UP000069272 UP000076408 UP000075883 UP000075901 UP000075882 UP000192221 UP000095301 UP000091820 UP000008792 UP000007798 UP000268350 UP000092443 UP000092460 UP000001819 UP000008711 UP000092553 UP000037069 UP000009192 UP000000304 UP000002282 UP000095300 UP000000803 UP000007801 UP000078200 UP000008744 UP000001292 UP000001070 UP000092445 UP000183832 UP000007875 UP000018468 UP000001646 UP000034805 UP000192224 UP000008672 UP000005225 UP000007303 UP000233140 UP000005207 UP000248482 UP000261460 UP000005226 UP000000715 UP000261420 UP000261540 UP000265100 UP000265160 UP000009136 UP000261520 UP000002281 UP000189705 UP000233060 UP000233180 UP000233200 UP000233120 UP000233100 UP000028761 UP000029965 UP000009130 UP000233080
PRIDE
Pfam
PF00110 wnt
ProteinModelPortal
A0A2H1V5X9
A0A2A4JWX0
A0A212EY09
A0A0L7KV16
A0A182J4L2
A0A182HRY4
+ More
Q17NQ4 A0A182XN77 A0A084WU42 Q16S11 A0A182V617 A0A182GZF1 A0A182QGS0 Q7PM75 A0A182K4F2 A0A182WGH8 A0A182P4Q9 A0A182NBQ3 A0A182TXB1 A0A182R3S5 W5JGY4 B0WGM3 A0A182F0Y2 A0A182YRM0 A0A182MG79 A0A182T9X4 A0A182KU44 A0A1W4VP50 A0A1W4VAJ5 A0A1I8MBQ3 A0A1A9WUF1 A0A1W4VBW8 B4LS45 B4MZW5 A0A3B0KEL3 A0A1A9YDM9 A0A1B0BKX4 A0A0R3NX81 B3N6A9 A0A0M3QTF8 A0A0L0BQQ3 A0A0R3NRN4 B4KKD1 B4Q5E0 B4NZM3 A0A1I8NSI0 A0A0J9QXL2 D2NUH8 P40589 B3MK95 A0A3B0JI52 A0A1A9V1H2 B4G848 B5DHZ8 B4HY45 B4JQN1 A0A0K8U2M1 A0A1A9Z613 W8C1N2 A0A0G3FA69 A0A0A1X1W6 A0A1J1I182 H2Y6S0 W5N397 G1KSZ4 A0A0P7WIA1 H3A9K0 H0WTI3 H3D297 A0A2K5XIG2 Q4S992 I3IYD6 A0A2Y9KWZ8 A0A3B4FPD0 H2Y6R9 H2UHV8 M3YA59 A0A3B4V283 A0A3B3RN63 A0A3P8NMX0 A0A3P9D6W4 F1N7G3 A0A3B3Z8A1 F7APE5 A0A3Q0FYK0 A0A2K5P6W2 A0A2K6LUB5 A0A2K6PMR6 A0A2K6CMG5 A0A2K5TKC3 A0A2I3N5M7 A0A2K5P6S0 A0A2K6CMG2 A0A096P675 A0A3B3RPE7 A0A0D9QUS2 G7PV23 G7NJ57 A0A2K5HBR8
Q17NQ4 A0A182XN77 A0A084WU42 Q16S11 A0A182V617 A0A182GZF1 A0A182QGS0 Q7PM75 A0A182K4F2 A0A182WGH8 A0A182P4Q9 A0A182NBQ3 A0A182TXB1 A0A182R3S5 W5JGY4 B0WGM3 A0A182F0Y2 A0A182YRM0 A0A182MG79 A0A182T9X4 A0A182KU44 A0A1W4VP50 A0A1W4VAJ5 A0A1I8MBQ3 A0A1A9WUF1 A0A1W4VBW8 B4LS45 B4MZW5 A0A3B0KEL3 A0A1A9YDM9 A0A1B0BKX4 A0A0R3NX81 B3N6A9 A0A0M3QTF8 A0A0L0BQQ3 A0A0R3NRN4 B4KKD1 B4Q5E0 B4NZM3 A0A1I8NSI0 A0A0J9QXL2 D2NUH8 P40589 B3MK95 A0A3B0JI52 A0A1A9V1H2 B4G848 B5DHZ8 B4HY45 B4JQN1 A0A0K8U2M1 A0A1A9Z613 W8C1N2 A0A0G3FA69 A0A0A1X1W6 A0A1J1I182 H2Y6S0 W5N397 G1KSZ4 A0A0P7WIA1 H3A9K0 H0WTI3 H3D297 A0A2K5XIG2 Q4S992 I3IYD6 A0A2Y9KWZ8 A0A3B4FPD0 H2Y6R9 H2UHV8 M3YA59 A0A3B4V283 A0A3B3RN63 A0A3P8NMX0 A0A3P9D6W4 F1N7G3 A0A3B3Z8A1 F7APE5 A0A3Q0FYK0 A0A2K5P6W2 A0A2K6LUB5 A0A2K6PMR6 A0A2K6CMG5 A0A2K5TKC3 A0A2I3N5M7 A0A2K5P6S0 A0A2K6CMG2 A0A096P675 A0A3B3RPE7 A0A0D9QUS2 G7PV23 G7NJ57 A0A2K5HBR8
PDB
6AHY
E-value=3.48911e-32,
Score=345
Ontologies
PATHWAY
GO
GO:0005102
GO:0005576
GO:0007275
GO:0016021
GO:0016055
GO:0030182
GO:0005109
GO:0045165
GO:0005615
GO:0016201
GO:0035567
GO:0009986
GO:0031290
GO:0001736
GO:0035206
GO:0008585
GO:0008045
GO:0016477
GO:0007435
GO:0003407
GO:0072046
GO:0060071
GO:1902455
GO:0035150
GO:0072174
GO:0030539
GO:0003339
GO:0072003
GO:0048018
GO:0001932
GO:0009267
GO:0039706
GO:0009786
GO:0072038
GO:0060021
GO:0048701
GO:0072181
GO:0043085
GO:0001658
GO:1905438
GO:0060070
GO:0061038
GO:0072044
GO:0001701
GO:0016020
GO:0005215
GO:0006810
GO:0006397
GO:0004651
GO:0005634
PANTHER
Topology
Subcellular location
SignalP
Position: 1 - 25,
Likelihood: 0.970625
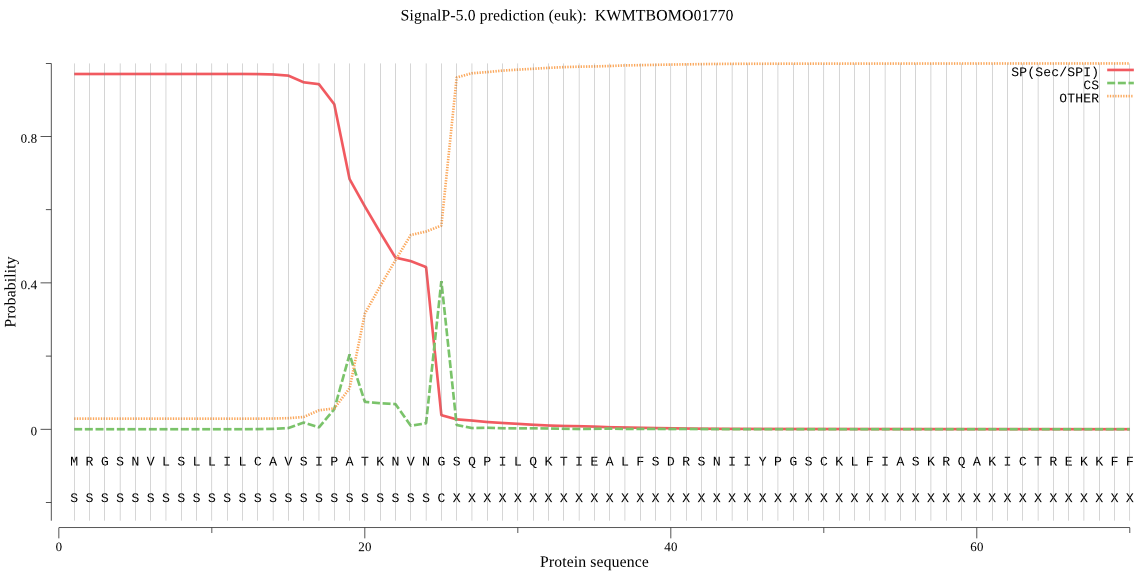
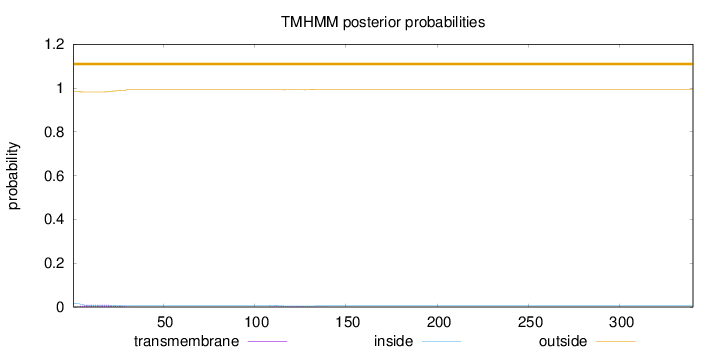
Length:
340
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26935
Exp number, first 60 AAs:
0.20502
Total prob of N-in:
0.01709
outside
1 - 340
Population Genetic Test Statistics
Pi
206.224614
Theta
196.05031
Tajima's D
0.051458
CLR
0
CSRT
0.385330733463327
Interpretation
Uncertain
