Gene
KWMTBOMO01767
Pre Gene Modal
BGIBMGA006005
Annotation
PREDICTED:_sodium-driven_chloride_bicarbonate_exchanger_isoform_X1_[Papilio_machaon]
Full name
Anion exchange protein
Location in the cell
Extracellular Reliability : 2.071 Nuclear Reliability : 3.818
Sequence
CDS
ATGGAGACGAGAGCTGATGATGAGGAAGCCCCGACCGACCCAGCAGCCAGGAAACAAACGTATCACGATTACACAGAGCAAGATTTTGAAGGCCATCGAGCCCACACAGTGTTTGTGGGTGTACACGTGCCCGCCCGCAGACACTCCCACAGGAAACACAAACACCACCACCGCAACGGGGAGAAGGAGAGCGAGAAACCAGGGCCCCCGATAATGTCTCGCGTCAGAAGATTGAGCGTTACGGACGACGGAGAAACATTGACTCCACCAGCACAGAGAGTGCAGTTCATACTTGGCGAGGATGTCGATGATAGCACACTTGAATCTCATCCTTTATTTTCCGAAATGGAAGAATTGGTCAAAGAGGGTGACGAACTGGAATGGAAAGAGACAGCTCGATGGATCAAATTCGAAGAAGACGTCGAAGAAGGGGGCAATCGTTGGTCTAAGCCACACGTTGCCACTTTATCATTGCATTCGCTTTTCGAACTCCGCAGCCTGATCCTAAACGGCTCTGTTATACTGGATATGGAAGCTAGTTCACTAGAACAGGTAGCGGATAATGTATTGAACAGCATGGTGAACGATGGGTCGCTTGGATATGACTCTAGAGAAAAAGTTAAGGATGCACTGCTAAGACGTCACAGACATCAACACGAAAGACGAAACCATAACATGTCTAGGCTACCTCTAATTAGATCGCTGGCCGACATTGGCCGCAATCACTCCTCTTGTAAAAATTTTCAGGATGGTGGCTCTCGACGCTCCAGTTCGAGGAGTTGTCGAGGGTCCACCTCATCACCTCACACGGCTCTACTCCAGGCAGCCGACGAAAGAGGCTGCTACCTAGCTGTTCCAGTGGAAGAAGGCGGTACAGGAAGTGGAAATTCTCACAAAGGAGAATTCAGTCGTTTCCTAGGCATCCCTAACGCGGGTGGTGCCAGCGACGAAGGCATGGTAAAGAGTCCAAGCAGCGTTTCAATGAATCGCCATCAGAGTACAGGCGATCTCCACTTGAACGGAGATATAAATCATAAGTGTAATTTAAATTTTATGAGAAAAATTCCACCAGGAGCGGAGGCCTCCAATATCTTAGTTGGAGAAGTCGATTTCCTTGAAAAAACATTATCAGCATTCGTAAGACTTAAAACCGGGACCATCATGGGTGATTTAACAGAAGTACCAGTCCCGACCAGGTTTATGTTCGTTCTATTAGGACCACCAAACAGCAATTCGAGTTTTCATGAAATCGGCCGTGCTATGGCCACATTGATGTCCGATGAAATATTTCACGAAGTCGCTTACCGAGCTAAAAAGAGGGATCATTTATTAGCTGGCATAGATGAATTTCTCGATGCTGTCACAGTTTTACCACCAGGTGAGTGGGACCCGGCCATCCGTATAGAACCACCAGCAGCTATACCCTCGCAAGATCCTCGGAAACGTCCTAATGATAATCCTAAAGATGAATTAGACGAAGAAGAAGAGGAGCAAAAGCAAAGAGAAGAGTCAGGATTAGCAAGAACAGGACGATTATTCGGTGGCCTAATAAATGATCTTAAAAGAAAAATGCCATGGTATTGGTCGGACTTCAAAGACGCTTTAGCTCTACAATGCATTGCCTCGTGGATATTTTTGTATTTTGCATGTCTATCACCCATAATAACTTTTGGCGGACTACTTGGCGAGGCGACTGGAAAGAATATGGCAGCAATGGAGTCTTTGGTATCGGGATTTGTCTGTGGAATGGGCTATGGTTTCTTTTCCGGACAACCGTTAACGATTTTGGGCTCGACGGGGCCCGTATTGGTTTTTGAAACGATTGTATTTGAATTTTGCAGACAAATAGGTTGGAATTATATGTCGTTTAGATTTTGCATTGGGACGTGGATCACGGTTATTTTGATCATATTGGTCGCGATCGACGCAAGTGCGCTTGTTTGTTACATAACAAGATTTACAGAGGAAAATTTTGCCACATTAATAGCCTTCATTTTCATTTATAAGGCAGTTGAAAACGTTATATCTATAGGAAAGAAATATCCTATGAAGACACGCGCCGACGAAGGCTTCAATTACGAGTGCTATTGTTTACCAACCAATTATTCTAGAGTACCGGAGCAATTTAATTGGTCCACTGTTGATAAGGAGTCTTGTCAGTTAAACAATGGAACGCTAACGGGAGAAGGATGCCATACAAAGGAATATGTACCAGACGTTTTTCTGATGTCCATAATACTGTTTCTAGGAACGTTCACAATTTCAATTATATTAAAGGACTTTAAAAACTCACTATTCTTTCCATCGAAGATAAGACAGGTCATAAGCGACTTTTCAGTTATTATTGCTATATTATCAATGTCATTCTTGGATTATAAAGTTGGGGTCAAAACTCCCAAACTCGAAGTACCATCCGAATTAAAGCCAACGTTGCCCACGAGAGATTGGGTTATTACACCATTCAATGGAAACCCGATATGGTCAGCCTTGGCAGCCTTACTGCCCGCTCTATTGGGCACCATACTTATTTTCATGGACCAACAAATAACTGCTGTTATCGTGAATCGTAAAGAGAACAAACTGAAGAAAGGCTGCGGCTACCATTTGGATTTATTCGTTTTGGCCATATTAATACAAATCTGTTCCGTTATGGGTTTGCCGTGGTTCGTAGCGGCCACGGTTCTTAGCATTAATCATGTGAATTCATTGAAAGTTGAATCGGAGTGTGCGGCTCCCGGAGAAAAGCCGCAATTTCTAGGAGTCAGAGAGCAAAGAGTTACCCATATACTGATTTTCTTGACCATAGGCTGCTCGGTTGTATTGACACCGGTTTTAAGGAACATTCCCATGCCGGTACTTTTTGGAGTCTTCTTGTATATGGGTGTAGCGTCACTAAAAGGCTTACAATTTTTCGATCGGATTCTGATAATGTTTATGCCTCAAAAATACCAGCCAGACCACATGTTTTTAAGACAGGTGCCTATACGACGCGTCCATCTTTTTACCGCAGTACAATTAACCTGCCTCGTTTGCTTGTGGGTAATAAAATCTTTCTCTCTGACGTCGATTTTATTCCCTTTAATGTTGGTCGTTATGATTGGAATAAGAAAGTCATTAGACTGCTTCTTTACGAGGAGGGAACTGAAAATACTGGACGACGTGATGCCGGAAATGACAAAGCGAAATCAGGACGAGCTGCGAGAACTTGGCGATGCTGAGGAATCAAATAAGAGCAATCCTCCAGCATACCAAGAAAACCTACAGATACCACTTATTAATGGAAATATTATGAACATACCGCTCACCTCTATCAATATCTCAGAGGAGGTAAACAAAACTGGTATATGGAAACAAGTTAATAGCAATAATAAGAGTCGTAACGATCATAAGAATAAATCCGGAAAGAAAAATAATAAACACAAAAAAAATATGTCGAAAAATGCACAAACGGAAATCGAAAGGAGACTGTCCGTAATGGACGAAGTCGAAGAAGACGATCCATCGGTTAAGAGCGATATTGTAAGGCGCAAGGACTCGTGGAGCGGTGGAATCAAAAAGAGTGAATCTGGAAAAAATAATACGTCTGCCGAAACCTCGGTGTAA
Protein
METRADDEEAPTDPAARKQTYHDYTEQDFEGHRAHTVFVGVHVPARRHSHRKHKHHHRNGEKESEKPGPPIMSRVRRLSVTDDGETLTPPAQRVQFILGEDVDDSTLESHPLFSEMEELVKEGDELEWKETARWIKFEEDVEEGGNRWSKPHVATLSLHSLFELRSLILNGSVILDMEASSLEQVADNVLNSMVNDGSLGYDSREKVKDALLRRHRHQHERRNHNMSRLPLIRSLADIGRNHSSCKNFQDGGSRRSSSRSCRGSTSSPHTALLQAADERGCYLAVPVEEGGTGSGNSHKGEFSRFLGIPNAGGASDEGMVKSPSSVSMNRHQSTGDLHLNGDINHKCNLNFMRKIPPGAEASNILVGEVDFLEKTLSAFVRLKTGTIMGDLTEVPVPTRFMFVLLGPPNSNSSFHEIGRAMATLMSDEIFHEVAYRAKKRDHLLAGIDEFLDAVTVLPPGEWDPAIRIEPPAAIPSQDPRKRPNDNPKDELDEEEEEQKQREESGLARTGRLFGGLINDLKRKMPWYWSDFKDALALQCIASWIFLYFACLSPIITFGGLLGEATGKNMAAMESLVSGFVCGMGYGFFSGQPLTILGSTGPVLVFETIVFEFCRQIGWNYMSFRFCIGTWITVILIILVAIDASALVCYITRFTEENFATLIAFIFIYKAVENVISIGKKYPMKTRADEGFNYECYCLPTNYSRVPEQFNWSTVDKESCQLNNGTLTGEGCHTKEYVPDVFLMSIILFLGTFTISIILKDFKNSLFFPSKIRQVISDFSVIIAILSMSFLDYKVGVKTPKLEVPSELKPTLPTRDWVITPFNGNPIWSALAALLPALLGTILIFMDQQITAVIVNRKENKLKKGCGYHLDLFVLAILIQICSVMGLPWFVAATVLSINHVNSLKVESECAAPGEKPQFLGVREQRVTHILIFLTIGCSVVLTPVLRNIPMPVLFGVFLYMGVASLKGLQFFDRILIMFMPQKYQPDHMFLRQVPIRRVHLFTAVQLTCLVCLWVIKSFSLTSILFPLMLVVMIGIRKSLDCFFTRRELKILDDVMPEMTKRNQDELRELGDAEESNKSNPPAYQENLQIPLINGNIMNIPLTSINISEEVNKTGIWKQVNSNNKSRNDHKNKSGKKNNKHKKNMSKNAQTEIERRLSVMDEVEEDDPSVKSDIVRRKDSWSGGIKKSESGKNNTSAETSV
Summary
Similarity
Belongs to the anion exchanger (TC 2.A.31) family.
Uniprot
A0A2A4JJY1
A0A212F572
A0A2H1WET2
A0A194R2X3
A0A0L7L825
H9J913
+ More
A0A087ZTR6 F4WZ58 A0A158NDP7 E2A0M0 A0A158NDP8 A0A154PGQ4 A0A3L8DZJ4 E2B606 A0A195DA49 A0A195EYK7 A0A0M8ZWA1 A0A026W453 A0A232ES63 K7JA63 A0A023EZ90 A0A224XDH8 A0A1B6GCA1 A0A0V0G4B1 G1UJV5 A0A1S4FRH4 Q16S12 Q16S13 A0A1S4JH65 A0A1B6H4L3 A0A067QYV1 C5J0I4 A0A182R3S6 A0A1I8QD05 A0A1I8QCT0 A0A1I8QCV8 A0A182QVR5 A0A1B0CVR6 A0A1I8QCW3 A0A1I8QCZ9 A0A1I8QCU6 C5H4P2 A0A182F0Y1 A0A182KU45 A0A1I8QCU4 A0A182V3P4 A0A182XN78 A0A0C9RDR5 A0A1I8QCV4 A0A1I8QCV1 A0A182Y0M6 A0A182P4Q8 A0A1S4JF98 A0A1S4JF84 A0A023EXA3 B0WGM2 A0A182J5W4 A0A1Q3FQD9 A0A139WI57 A0A084WU40 A0A182NBQ4 A0A0R3NS80 A0A1Q3FQP1 A0A1Q3FQI0 Q7Q1L0 G1UJV4 A0A0Q9WAS4 A0A0Q9X5X7 Q6WLH7 A0A0K8SV49 A0A0A9WIM9 A0A0A9WLI8 A0A0K8SVL4 A0A0K8SWB4 A0A0K8SV55 A0A0Q9X7W1 A0A0Q9XI36 A0A0J9QX90 A0A0J9QXJ0 A0A0J9QXL4 A0A0Q9X5F1 A0A0P8XGC1 A0A0P8XGU8 A0A2M4BB20 A0A0R3NYG5 A0A0Q5WNF1 A0A0Q9XGV3 A0A0R3NRW7 A0A0R3NS27 A0A0Q9WJL6 A0A0Q9WBI2 A0A0J9THR5 A0A0Q9WL83 A0A0Q9WLY6 A0A0P8Y4N8 A0A0Q9XFE9 A0A0R1DJP8 A0A0Q9WAX6 A0A0R1DJH0 A0A0R3NSF8
A0A087ZTR6 F4WZ58 A0A158NDP7 E2A0M0 A0A158NDP8 A0A154PGQ4 A0A3L8DZJ4 E2B606 A0A195DA49 A0A195EYK7 A0A0M8ZWA1 A0A026W453 A0A232ES63 K7JA63 A0A023EZ90 A0A224XDH8 A0A1B6GCA1 A0A0V0G4B1 G1UJV5 A0A1S4FRH4 Q16S12 Q16S13 A0A1S4JH65 A0A1B6H4L3 A0A067QYV1 C5J0I4 A0A182R3S6 A0A1I8QD05 A0A1I8QCT0 A0A1I8QCV8 A0A182QVR5 A0A1B0CVR6 A0A1I8QCW3 A0A1I8QCZ9 A0A1I8QCU6 C5H4P2 A0A182F0Y1 A0A182KU45 A0A1I8QCU4 A0A182V3P4 A0A182XN78 A0A0C9RDR5 A0A1I8QCV4 A0A1I8QCV1 A0A182Y0M6 A0A182P4Q8 A0A1S4JF98 A0A1S4JF84 A0A023EXA3 B0WGM2 A0A182J5W4 A0A1Q3FQD9 A0A139WI57 A0A084WU40 A0A182NBQ4 A0A0R3NS80 A0A1Q3FQP1 A0A1Q3FQI0 Q7Q1L0 G1UJV4 A0A0Q9WAS4 A0A0Q9X5X7 Q6WLH7 A0A0K8SV49 A0A0A9WIM9 A0A0A9WLI8 A0A0K8SVL4 A0A0K8SWB4 A0A0K8SV55 A0A0Q9X7W1 A0A0Q9XI36 A0A0J9QX90 A0A0J9QXJ0 A0A0J9QXL4 A0A0Q9X5F1 A0A0P8XGC1 A0A0P8XGU8 A0A2M4BB20 A0A0R3NYG5 A0A0Q5WNF1 A0A0Q9XGV3 A0A0R3NRW7 A0A0R3NS27 A0A0Q9WJL6 A0A0Q9WBI2 A0A0J9THR5 A0A0Q9WL83 A0A0Q9WLY6 A0A0P8Y4N8 A0A0Q9XFE9 A0A0R1DJP8 A0A0Q9WAX6 A0A0R1DJH0 A0A0R3NSF8
Pubmed
EMBL
NWSH01001317
PCG71733.1
AGBW02010235
OWR48880.1
ODYU01008194
SOQ51588.1
+ More
KQ460855 KPJ11869.1 JTDY01002421 KOB71446.1 BABH01004706 BABH01004707 BABH01004708 BABH01004709 GL888465 EGI60561.1 ADTU01012762 ADTU01012763 ADTU01012764 GL435626 EFN72923.1 KQ434899 KZC10987.1 QOIP01000002 RLU25653.1 GL445900 EFN88867.1 KQ981082 KYN09741.1 KQ981906 KYN33303.1 KQ435851 KOX70859.1 KK107436 EZA50865.1 NNAY01002482 OXU21209.1 AAZX01001626 AAZX01012463 AAZX01013009 GBBI01004177 JAC14535.1 GFTR01008588 JAW07838.1 GECZ01009701 JAS60068.1 GECL01003257 JAP02867.1 AB669028 BAK74835.1 CH477689 EAT37255.1 EAT37256.1 GECZ01000147 JAS69622.1 KK853096 KDR11468.1 FJ981607 ACR56333.1 AXCN02000684 AJWK01031035 AJWK01031036 AJWK01031037 AJWK01031038 AJWK01031039 EU719196 ACH96582.1 GBYB01006475 JAG76242.1 GAPW01000057 JAC13541.1 DS231927 EDS27051.1 GFDL01005214 JAV29831.1 KQ971342 KYB27584.1 ATLV01026995 ATLV01026996 KE525421 KFB53734.1 CH379060 KRT03865.1 GFDL01005282 JAV29763.1 GFDL01005251 JAV29794.1 AAAB01008980 EAA14173.3 AB669027 BAK74834.1 CH940649 KRF81827.1 CH933807 KRG03357.1 AY280611 AAQ21364.1 GBRD01008633 JAG57188.1 GBHO01035307 GBHO01035306 JAG08297.1 JAG08298.1 GBHO01035308 GBHO01035304 JAG08296.1 JAG08300.1 GBRD01008634 GBRD01008632 GBRD01008630 JAG57191.1 GBRD01008631 GBRD01008629 GBRD01008627 JAG57190.1 GBRD01008628 JAG57193.1 KRG03369.1 KRG03368.1 CM002910 KMY88727.1 KMY88726.1 KMY88723.1 KRG03371.1 CH902620 KPU73960.1 KPU73970.1 GGFJ01001086 MBW50227.1 KRT03859.1 CH954177 KQS70682.1 KRG03367.1 KRT03858.1 KRT03860.1 KRF81825.1 KRF81826.1 KMY88735.1 KRF81828.1 KRF81835.1 KPU73962.1 KRG03358.1 CM000157 KRJ97384.1 KRF81832.1 KRJ97388.1 KRT03868.1
KQ460855 KPJ11869.1 JTDY01002421 KOB71446.1 BABH01004706 BABH01004707 BABH01004708 BABH01004709 GL888465 EGI60561.1 ADTU01012762 ADTU01012763 ADTU01012764 GL435626 EFN72923.1 KQ434899 KZC10987.1 QOIP01000002 RLU25653.1 GL445900 EFN88867.1 KQ981082 KYN09741.1 KQ981906 KYN33303.1 KQ435851 KOX70859.1 KK107436 EZA50865.1 NNAY01002482 OXU21209.1 AAZX01001626 AAZX01012463 AAZX01013009 GBBI01004177 JAC14535.1 GFTR01008588 JAW07838.1 GECZ01009701 JAS60068.1 GECL01003257 JAP02867.1 AB669028 BAK74835.1 CH477689 EAT37255.1 EAT37256.1 GECZ01000147 JAS69622.1 KK853096 KDR11468.1 FJ981607 ACR56333.1 AXCN02000684 AJWK01031035 AJWK01031036 AJWK01031037 AJWK01031038 AJWK01031039 EU719196 ACH96582.1 GBYB01006475 JAG76242.1 GAPW01000057 JAC13541.1 DS231927 EDS27051.1 GFDL01005214 JAV29831.1 KQ971342 KYB27584.1 ATLV01026995 ATLV01026996 KE525421 KFB53734.1 CH379060 KRT03865.1 GFDL01005282 JAV29763.1 GFDL01005251 JAV29794.1 AAAB01008980 EAA14173.3 AB669027 BAK74834.1 CH940649 KRF81827.1 CH933807 KRG03357.1 AY280611 AAQ21364.1 GBRD01008633 JAG57188.1 GBHO01035307 GBHO01035306 JAG08297.1 JAG08298.1 GBHO01035308 GBHO01035304 JAG08296.1 JAG08300.1 GBRD01008634 GBRD01008632 GBRD01008630 JAG57191.1 GBRD01008631 GBRD01008629 GBRD01008627 JAG57190.1 GBRD01008628 JAG57193.1 KRG03369.1 KRG03368.1 CM002910 KMY88727.1 KMY88726.1 KMY88723.1 KRG03371.1 CH902620 KPU73960.1 KPU73970.1 GGFJ01001086 MBW50227.1 KRT03859.1 CH954177 KQS70682.1 KRG03367.1 KRT03858.1 KRT03860.1 KRF81825.1 KRF81826.1 KMY88735.1 KRF81828.1 KRF81835.1 KPU73962.1 KRG03358.1 CM000157 KRJ97384.1 KRF81832.1 KRJ97388.1 KRT03868.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000037510
UP000005204
UP000005203
+ More
UP000007755 UP000005205 UP000000311 UP000076502 UP000279307 UP000008237 UP000078492 UP000078541 UP000053105 UP000053097 UP000215335 UP000002358 UP000008820 UP000027135 UP000075900 UP000095300 UP000075886 UP000092461 UP000069272 UP000075882 UP000075903 UP000076407 UP000076408 UP000075885 UP000002320 UP000075880 UP000007266 UP000030765 UP000075884 UP000001819 UP000007062 UP000008792 UP000009192 UP000007801 UP000008711 UP000002282
UP000007755 UP000005205 UP000000311 UP000076502 UP000279307 UP000008237 UP000078492 UP000078541 UP000053105 UP000053097 UP000215335 UP000002358 UP000008820 UP000027135 UP000075900 UP000095300 UP000075886 UP000092461 UP000069272 UP000075882 UP000075903 UP000076407 UP000076408 UP000075885 UP000002320 UP000075880 UP000007266 UP000030765 UP000075884 UP000001819 UP000007062 UP000008792 UP000009192 UP000007801 UP000008711 UP000002282
PRIDE
Interpro
SUPFAM
SSF55804
SSF55804
Gene 3D
ProteinModelPortal
A0A2A4JJY1
A0A212F572
A0A2H1WET2
A0A194R2X3
A0A0L7L825
H9J913
+ More
A0A087ZTR6 F4WZ58 A0A158NDP7 E2A0M0 A0A158NDP8 A0A154PGQ4 A0A3L8DZJ4 E2B606 A0A195DA49 A0A195EYK7 A0A0M8ZWA1 A0A026W453 A0A232ES63 K7JA63 A0A023EZ90 A0A224XDH8 A0A1B6GCA1 A0A0V0G4B1 G1UJV5 A0A1S4FRH4 Q16S12 Q16S13 A0A1S4JH65 A0A1B6H4L3 A0A067QYV1 C5J0I4 A0A182R3S6 A0A1I8QD05 A0A1I8QCT0 A0A1I8QCV8 A0A182QVR5 A0A1B0CVR6 A0A1I8QCW3 A0A1I8QCZ9 A0A1I8QCU6 C5H4P2 A0A182F0Y1 A0A182KU45 A0A1I8QCU4 A0A182V3P4 A0A182XN78 A0A0C9RDR5 A0A1I8QCV4 A0A1I8QCV1 A0A182Y0M6 A0A182P4Q8 A0A1S4JF98 A0A1S4JF84 A0A023EXA3 B0WGM2 A0A182J5W4 A0A1Q3FQD9 A0A139WI57 A0A084WU40 A0A182NBQ4 A0A0R3NS80 A0A1Q3FQP1 A0A1Q3FQI0 Q7Q1L0 G1UJV4 A0A0Q9WAS4 A0A0Q9X5X7 Q6WLH7 A0A0K8SV49 A0A0A9WIM9 A0A0A9WLI8 A0A0K8SVL4 A0A0K8SWB4 A0A0K8SV55 A0A0Q9X7W1 A0A0Q9XI36 A0A0J9QX90 A0A0J9QXJ0 A0A0J9QXL4 A0A0Q9X5F1 A0A0P8XGC1 A0A0P8XGU8 A0A2M4BB20 A0A0R3NYG5 A0A0Q5WNF1 A0A0Q9XGV3 A0A0R3NRW7 A0A0R3NS27 A0A0Q9WJL6 A0A0Q9WBI2 A0A0J9THR5 A0A0Q9WL83 A0A0Q9WLY6 A0A0P8Y4N8 A0A0Q9XFE9 A0A0R1DJP8 A0A0Q9WAX6 A0A0R1DJH0 A0A0R3NSF8
A0A087ZTR6 F4WZ58 A0A158NDP7 E2A0M0 A0A158NDP8 A0A154PGQ4 A0A3L8DZJ4 E2B606 A0A195DA49 A0A195EYK7 A0A0M8ZWA1 A0A026W453 A0A232ES63 K7JA63 A0A023EZ90 A0A224XDH8 A0A1B6GCA1 A0A0V0G4B1 G1UJV5 A0A1S4FRH4 Q16S12 Q16S13 A0A1S4JH65 A0A1B6H4L3 A0A067QYV1 C5J0I4 A0A182R3S6 A0A1I8QD05 A0A1I8QCT0 A0A1I8QCV8 A0A182QVR5 A0A1B0CVR6 A0A1I8QCW3 A0A1I8QCZ9 A0A1I8QCU6 C5H4P2 A0A182F0Y1 A0A182KU45 A0A1I8QCU4 A0A182V3P4 A0A182XN78 A0A0C9RDR5 A0A1I8QCV4 A0A1I8QCV1 A0A182Y0M6 A0A182P4Q8 A0A1S4JF98 A0A1S4JF84 A0A023EXA3 B0WGM2 A0A182J5W4 A0A1Q3FQD9 A0A139WI57 A0A084WU40 A0A182NBQ4 A0A0R3NS80 A0A1Q3FQP1 A0A1Q3FQI0 Q7Q1L0 G1UJV4 A0A0Q9WAS4 A0A0Q9X5X7 Q6WLH7 A0A0K8SV49 A0A0A9WIM9 A0A0A9WLI8 A0A0K8SVL4 A0A0K8SWB4 A0A0K8SV55 A0A0Q9X7W1 A0A0Q9XI36 A0A0J9QX90 A0A0J9QXJ0 A0A0J9QXL4 A0A0Q9X5F1 A0A0P8XGC1 A0A0P8XGU8 A0A2M4BB20 A0A0R3NYG5 A0A0Q5WNF1 A0A0Q9XGV3 A0A0R3NRW7 A0A0R3NS27 A0A0Q9WJL6 A0A0Q9WBI2 A0A0J9THR5 A0A0Q9WL83 A0A0Q9WLY6 A0A0P8Y4N8 A0A0Q9XFE9 A0A0R1DJP8 A0A0Q9WAX6 A0A0R1DJH0 A0A0R3NSF8
PDB
6CAA
E-value=0,
Score=1955
Ontologies
GO
PANTHER
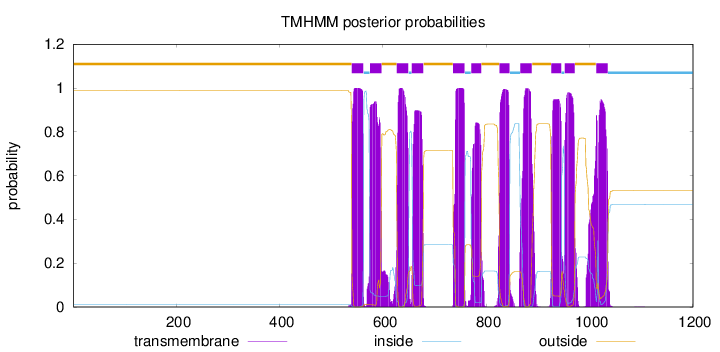
Topology
Subcellular location
Membrane
Length:
1200
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
237.4627
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.01070
outside
1 - 539
TMhelix
540 - 562
inside
563 - 574
TMhelix
575 - 597
outside
598 - 626
TMhelix
627 - 649
inside
650 - 655
TMhelix
656 - 678
outside
679 - 735
TMhelix
736 - 758
inside
759 - 770
TMhelix
771 - 790
outside
791 - 825
TMhelix
826 - 845
inside
846 - 865
TMhelix
866 - 888
outside
889 - 925
TMhelix
926 - 945
inside
946 - 951
TMhelix
952 - 971
outside
972 - 1012
TMhelix
1013 - 1035
inside
1036 - 1200
Population Genetic Test Statistics
Pi
211.240249
Theta
157.295706
Tajima's D
1.024953
CLR
0.974816
CSRT
0.665116744162792
Interpretation
Uncertain
