Gene
KWMTBOMO01766 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006144
Annotation
PREDICTED:_elongation_factor_G?_mitochondrial_[Amyelois_transitella]
Full name
Elongation factor G, mitochondrial
Alternative Name
Elongation factor G 1, mitochondrial
Elongation factor G1
Protein iconoclast
Elongation factor G1
Protein iconoclast
Location in the cell
Cytoplasmic Reliability : 3.148
Sequence
CDS
ATGACAATTTCTTATGCATTTCGTTTAATAAATGCGCTTAAAAATACAAAAAAATCGAGTGTATTTCAAATTGAGTTACAGAAATATTCATCACATGTCAAATATGCAGAACACAAACCATTGGAAAATATTCGAAATATTGGTATATCAGCTCACATAGACTCTGGAAAGACAACACTCACTGAACGTATTTTGTTCTATACTGGACGAATAGACCAAATGCACGAGGTTAAAGGTAAAGACAATGTTGGTGCTGTTATGGACTCAATGGAGTTGGAACGTCAAAGAGGAATTACAATACAGTCTGCTGCAACATACACTATTTGGAAGGAACATAACATTAACATTATTGATACTCCAGGACATGTAGATTTCACAGTAGAAGTAGAAAGAGCATTAAGAGTTTTAGATGGGGCGATATTGGTGTTGTGTTCTGTAGGTGGTGTTCAGAGCCAAACTTTAACAGTCAACAGACAAATGAAACGGTATAATGTACCATGTCTTGCATTTATCAATAAACTGGACAGATTAGGAGCTAATCCAGAAAGAGTACTGAAACAAATGCGTTCAAAATTAAAACATAATGCTGCATTTATACATCTACCAATTGGCTTAGAAAAAGAGTGTCAAGGAATTTTGGACCTTGTTGAGGAGAAAGCTATATATTTTGATGGTGAATATGGCGAGCATGTGCGGCTCGATGAGATTCCACAAGATCGTAGAACCGAAGTTAAGGAAAGGAGACATGAATTGATTGAACATCTTTCAAATGTAGATGAAACATTGGGAGAGTTATTCCTAGAAGAGAAAACTCCGACAGTCAATGATCTCAAGCAAGCATTACGGCGTTCCACATTAAAACGAGCATTCACTCCAGTGCTTCTGGGTACAGCTCTAAAGAATAAGGGAATTCAACCCTTACTGGATGCTGTGCTCACATATCTACCGCATCCAGGAGAAGTAGAAAACACTGCCTTGTTAAATGAAAGCAAAGATGGTGAAGGCCAAACCATAGTGTTGGATCCGACTAGGGATAATAAGAAACCATTTGTTGGTTTGGCATTCAAGTTGGAAGTGAGCAAATTTGGTCAACTCACATACTTGAGGTGTTATCAAGGGATGTTGAAGAGAGGTGACCATATATTCAACGCCAGGACTTACAAAAAGGTAAAAATATCACGTTTAGTCCGCATGCACTCGAACAACATGGAAGAGGTTAACGAGGTCCTGGCCGGTGATATATTCGCGTTGTTCGGTGTGGACTGCGCTTCGGGAGACACATTCGTGAACGATGCCAAACTGCAGCTCGCAATGGAGTCCATACATGTTCCTGACCCAGTCGTGTCCATGTCCATCAAACCGAAGCAAAATAAAGACAGAGATAACTTTTCTAAGGCTGTTGCTAGATTCACAAAAGAAGATCCCACTTTCCAATTTCATTACGACTCCGACAATAAGGAAACTATAGTGTCCGGTATGGGCGAACTCCATCTTGAGATCTATGCTCAAAGAATGGAGAGAGAATACAACTGTCCGGTCGAACTTGGGAAACCCAAGGTTGCTTTCAGGGAGACGCTGATTTCGCCTTGCGCTTTCGACTATTTACATAAGAAACAGTCGGGCGGAGCTGGCCAGTACGCACGAGTCAGCGGCATCATAGAACCTTTACCTCCTCACCAAAACACAGTACTCGAATTCGTTGATGAAACTGTGGGTACGAACATTCCCAAGCAGTTCGTGCCGGGTGTTGAAAGAGGTTTCCTGGATACATGCGAGAAGGGCTACCTGTCCGGGCACAAGGTCTCCGGTGTGAAGTTCAGGCTTCAGGACGGAGCCCACCACATTGTGGACTCCAGTGAGCTGGCATTCTTCTTGGCAGCTAAGGGGGCCGTCAAGGATGTTTTCGACGTGGGAGCGTGGCAGATACTCGAACCGGTCATGTTCGTTGAAGTAACGTTGCCGGACGAGTTCCACGGCACGGTGGTCGGGCAGCTCAACAAACGCGGCGGCATCATCACCGGGACCGAGGGGGCCGAGGGCTGGACCTCCATCTACGCCGAGGTTCCTCTCAACAACATGTTCGGATACGCCGGAGAACTGAGGTCAATGACTCAAGGTAAAGGAGAGTTCAGTATGGAGTACAGCCGGTACTCGCCTTGTCTACCGGACGTGCAGGAGCAACTGATCAGGAAGCACCAAGAGGAAATGGGGCTGCTACCAGACCAGAAGAAAAAGAAAAATTAA
Protein
MTISYAFRLINALKNTKKSSVFQIELQKYSSHVKYAEHKPLENIRNIGISAHIDSGKTTLTERILFYTGRIDQMHEVKGKDNVGAVMDSMELERQRGITIQSAATYTIWKEHNINIIDTPGHVDFTVEVERALRVLDGAILVLCSVGGVQSQTLTVNRQMKRYNVPCLAFINKLDRLGANPERVLKQMRSKLKHNAAFIHLPIGLEKECQGILDLVEEKAIYFDGEYGEHVRLDEIPQDRRTEVKERRHELIEHLSNVDETLGELFLEEKTPTVNDLKQALRRSTLKRAFTPVLLGTALKNKGIQPLLDAVLTYLPHPGEVENTALLNESKDGEGQTIVLDPTRDNKKPFVGLAFKLEVSKFGQLTYLRCYQGMLKRGDHIFNARTYKKVKISRLVRMHSNNMEEVNEVLAGDIFALFGVDCASGDTFVNDAKLQLAMESIHVPDPVVSMSIKPKQNKDRDNFSKAVARFTKEDPTFQFHYDSDNKETIVSGMGELHLEIYAQRMEREYNCPVELGKPKVAFRETLISPCAFDYLHKKQSGGAGQYARVSGIIEPLPPHQNTVLEFVDETVGTNIPKQFVPGVERGFLDTCEKGYLSGHKVSGVKFRLQDGAHHIVDSSELAFFLAAKGAVKDVFDVGAWQILEPVMFVEVTLPDEFHGTVVGQLNKRGGIITGTEGAEGWTSIYAEVPLNNMFGYAGELRSMTQGKGEFSMEYSRYSPCLPDVQEQLIRKHQEEMGLLPDQKKKKN
Summary
Description
Mitochondrial GTPase that catalyzes the GTP-dependent ribosomal translocation step during translation elongation. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the newly formed A-site-bound peptidyl-tRNA and P-site-bound deacylated tRNA move to the P and E sites, respectively. Catalyzes the coordinated movement of the two tRNA molecules, the mRNA and conformational changes in the ribosome.
Mitochondrial GTPase that catalyzes the GTP-dependent ribosomal translocation step during translation elongation. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the newly formed A-site-bound peptidyl-tRNA and P-site-bound deacylated tRNA move to the P and E sites, respectively. Catalyzes the coordinated movement of the two tRNA molecules, the mRNA and conformational changes in the ribosome. Essential during development as it acts as a retrograde signal from mitochondria to the nucleus to slow down cell proliferation if mitochondrial energy output is low (By similarity).
Mitochondrial GTPase that catalyzes the GTP-dependent ribosomal translocation step during translation elongation. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the newly formed A-site-bound peptidyl-tRNA and P-site-bound deacylated tRNA move to the P and E sites, respectively. Catalyzes the coordinated movement of the two tRNA molecules, the mRNA and conformational changes in the ribosome (By similarity). Essential during development as it acts as a retrograde signal from mitochondria to the nucleus to slow down cell proliferation if mitochondrial energy output is low.
Mitochondrial GTPase that catalyzes the GTP-dependent ribosomal translocation step during translation elongation. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the newly formed A-site-bound peptidyl-tRNA and P-site-bound deacylated tRNA move to the P and E sites, respectively. Catalyzes the coordinated movement of the two tRNA molecules, the mRNA and conformational changes in the ribosome. Essential during development as it acts as a retrograde signal from mitochondria to the nucleus to slow down cell proliferation if mitochondrial energy output is low (By similarity).
Mitochondrial GTPase that catalyzes the GTP-dependent ribosomal translocation step during translation elongation. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the newly formed A-site-bound peptidyl-tRNA and P-site-bound deacylated tRNA move to the P and E sites, respectively. Catalyzes the coordinated movement of the two tRNA molecules, the mRNA and conformational changes in the ribosome (By similarity). Essential during development as it acts as a retrograde signal from mitochondria to the nucleus to slow down cell proliferation if mitochondrial energy output is low.
Miscellaneous
This protein may be expected to contain an N-terminal transit peptide but none has been predicted.
Similarity
Belongs to the GTP-binding elongation factor family. EF-G/EF-2 subfamily.
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. EF-G/EF-2 subfamily.
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. EF-G/EF-2 subfamily.
Keywords
Complete proteome
Elongation factor
GTP-binding
Mitochondrion
Nucleotide-binding
Protein biosynthesis
Reference proteome
Transit peptide
Feature
chain Elongation factor G, mitochondrial
Uniprot
A0A2A4JK07
A0A194R253
A0A194PZ19
A0A212F564
A0A1Q3FEM1
B0WGM1
+ More
A0A336MWZ0 A0A182WGI1 A0A336KYM9 A0A1I8MS52 A0A1A9WUF6 A0A182GGX6 Q16S14 A0A023EVY5 A0A182M287 A0A182TUU2 A0A084WU39 A0A182HRY1 A0A182XN79 A0A182VJA8 A0A182KU31 A0A182P4Q7 Q7Q1K8 A0A2K7P6C4 A0A1A9YDN1 A0A182R3S7 A0A182NBQ5 A0A182Y0M7 A0A182F0X9 A0A1B0G1L1 A0A2H1X234 A0A0L0BS00 A0A1B0BKX1 A0A182K0G7 A0A182IX89 A0A1A9Z620 A0A1I8Q4C7 A0A182SQM6 A0A1A9V1H1 A0A182QYP8 W5JI53 A0A1J1I174 A0A2M4ADT9 A0A2M4BF04 T1DQ02 A0A2M4BF99 B4KKD5 A0A2M3Z4A2 A0A2M4BF03 A0A1W4UYC7 B4LS49 A0A1W4X6N8 B3N6A5 A0A0J9QX85 A0A034VEY2 A0A0M4EEV7 B4Q5D5 B4HY41 A0A0K8W112 U5EXB5 Q9VM33 B4NZM7 A0A0A1WI02 W8CD33 A0A1B0CWQ0 A0A3B0JMF7 B3MK91 D6WJJ3 B4MZW9 B4JQM7 A0A067QYF5 Q29N77 H9J9F1 A0A2J7RPP8 A0A1B6EF77 A0A0T6B5D7 A0A0L7L7V4 A0A1Y1M2G3 E2A0M4 A0A1L8DR80 K7JA58 A0A0J7L508 A0A195EZJ3 A0A232EEL7 A0A146LZS1 A0A1B6JPJ1 E9JBK2 F4W861 A0A0A9ZGU0 A0A348G619 A0A0K8T3P1 T1HPG2 A0A023F4E6 A0A0C9RR10 A0A195CLD9 A0A1B6G3A0 A0A0C9R809 A0A3L8E232 A0A195D8V1 A0A224X899
A0A336MWZ0 A0A182WGI1 A0A336KYM9 A0A1I8MS52 A0A1A9WUF6 A0A182GGX6 Q16S14 A0A023EVY5 A0A182M287 A0A182TUU2 A0A084WU39 A0A182HRY1 A0A182XN79 A0A182VJA8 A0A182KU31 A0A182P4Q7 Q7Q1K8 A0A2K7P6C4 A0A1A9YDN1 A0A182R3S7 A0A182NBQ5 A0A182Y0M7 A0A182F0X9 A0A1B0G1L1 A0A2H1X234 A0A0L0BS00 A0A1B0BKX1 A0A182K0G7 A0A182IX89 A0A1A9Z620 A0A1I8Q4C7 A0A182SQM6 A0A1A9V1H1 A0A182QYP8 W5JI53 A0A1J1I174 A0A2M4ADT9 A0A2M4BF04 T1DQ02 A0A2M4BF99 B4KKD5 A0A2M3Z4A2 A0A2M4BF03 A0A1W4UYC7 B4LS49 A0A1W4X6N8 B3N6A5 A0A0J9QX85 A0A034VEY2 A0A0M4EEV7 B4Q5D5 B4HY41 A0A0K8W112 U5EXB5 Q9VM33 B4NZM7 A0A0A1WI02 W8CD33 A0A1B0CWQ0 A0A3B0JMF7 B3MK91 D6WJJ3 B4MZW9 B4JQM7 A0A067QYF5 Q29N77 H9J9F1 A0A2J7RPP8 A0A1B6EF77 A0A0T6B5D7 A0A0L7L7V4 A0A1Y1M2G3 E2A0M4 A0A1L8DR80 K7JA58 A0A0J7L508 A0A195EZJ3 A0A232EEL7 A0A146LZS1 A0A1B6JPJ1 E9JBK2 F4W861 A0A0A9ZGU0 A0A348G619 A0A0K8T3P1 T1HPG2 A0A023F4E6 A0A0C9RR10 A0A195CLD9 A0A1B6G3A0 A0A0C9R809 A0A3L8E232 A0A195D8V1 A0A224X899
Pubmed
26354079
22118469
25315136
26483478
17510324
24945155
+ More
24438588 20966253 12364791 25244985 26108605 20920257 23761445 17994087 22936249 25348373 21364917 10731132 12537572 25830018 24495485 18362917 19820115 24845553 15632085 19121390 26227816 28004739 20798317 20075255 28648823 26823975 21282665 21719571 25401762 25474469 30249741
24438588 20966253 12364791 25244985 26108605 20920257 23761445 17994087 22936249 25348373 21364917 10731132 12537572 25830018 24495485 18362917 19820115 24845553 15632085 19121390 26227816 28004739 20798317 20075255 28648823 26823975 21282665 21719571 25401762 25474469 30249741
EMBL
NWSH01001317
PCG71732.1
KQ460855
KPJ11868.1
KQ459584
KPI98577.1
+ More
AGBW02010235 OWR48878.1 GFDL01009051 JAV25994.1 DS231927 UFQT01002506 SSX33599.1 UFQS01001105 UFQT01001105 SSX09025.1 SSX28936.1 JXUM01062623 KQ562208 KXJ76412.1 CH477689 GAPW01000311 JAC13287.1 AXCM01001134 ATLV01026995 KE525421 KFB53733.1 APCN01001775 AAAB01008980 CCAG010021325 ODYU01012882 SOQ59410.1 JRES01001454 KNC22817.1 JXJN01016076 AXCN02000684 ADMH02001427 ETN62580.1 CVRI01000037 CRK93506.1 GGFK01005630 MBW38951.1 GGFJ01002488 MBW51629.1 GAMD01002874 JAA98716.1 GGFJ01002490 MBW51631.1 CH933807 GGFM01002527 MBW23278.1 GGFJ01002489 MBW51630.1 CH940649 CH954177 CM002910 KMY88722.1 GAKP01017086 JAC41866.1 CP012523 ALC38805.1 CM000361 CH480818 GDHF01007550 JAI44764.1 GANO01001033 JAB58838.1 AE014134 BT023763 CM000157 GBXI01015623 JAC98668.1 GAMC01002646 JAC03910.1 AJWK01032753 OUUW01000006 SPP82033.1 CH902620 KQ971342 EFA04773.2 CH963920 CH916372 KK852829 KDR15371.1 CH379060 BABH01004703 BABH01004704 BABH01004705 NEVH01001358 PNF42811.1 GEDC01000784 JAS36514.1 LJIG01009669 KRT82618.1 JTDY01002421 KOB71445.1 GEZM01042339 JAV79929.1 GL435626 EFN72927.1 GFDF01005108 JAV08976.1 AAZX01001625 LBMM01000752 KMQ97569.1 KQ981906 KYN33299.1 NNAY01005274 OXU16800.1 GDHC01005538 JAQ13091.1 GECU01006538 JAT01169.1 GL771210 EFZ09794.1 GL887888 EGI69731.1 GBHO01000786 JAG42818.1 FX985558 BBF97892.1 GBRD01006016 JAG59805.1 ACPB03001467 GBBI01002326 JAC16386.1 GBYB01004168 GBYB01004169 GBYB01009716 JAG73935.1 JAG73936.1 JAG79483.1 KQ977600 KYN01518.1 GECZ01012986 JAS56783.1 GBYB01004170 JAG73937.1 QOIP01000002 RLU26078.1 KQ981153 KYN08854.1 GFTR01007850 JAW08576.1
AGBW02010235 OWR48878.1 GFDL01009051 JAV25994.1 DS231927 UFQT01002506 SSX33599.1 UFQS01001105 UFQT01001105 SSX09025.1 SSX28936.1 JXUM01062623 KQ562208 KXJ76412.1 CH477689 GAPW01000311 JAC13287.1 AXCM01001134 ATLV01026995 KE525421 KFB53733.1 APCN01001775 AAAB01008980 CCAG010021325 ODYU01012882 SOQ59410.1 JRES01001454 KNC22817.1 JXJN01016076 AXCN02000684 ADMH02001427 ETN62580.1 CVRI01000037 CRK93506.1 GGFK01005630 MBW38951.1 GGFJ01002488 MBW51629.1 GAMD01002874 JAA98716.1 GGFJ01002490 MBW51631.1 CH933807 GGFM01002527 MBW23278.1 GGFJ01002489 MBW51630.1 CH940649 CH954177 CM002910 KMY88722.1 GAKP01017086 JAC41866.1 CP012523 ALC38805.1 CM000361 CH480818 GDHF01007550 JAI44764.1 GANO01001033 JAB58838.1 AE014134 BT023763 CM000157 GBXI01015623 JAC98668.1 GAMC01002646 JAC03910.1 AJWK01032753 OUUW01000006 SPP82033.1 CH902620 KQ971342 EFA04773.2 CH963920 CH916372 KK852829 KDR15371.1 CH379060 BABH01004703 BABH01004704 BABH01004705 NEVH01001358 PNF42811.1 GEDC01000784 JAS36514.1 LJIG01009669 KRT82618.1 JTDY01002421 KOB71445.1 GEZM01042339 JAV79929.1 GL435626 EFN72927.1 GFDF01005108 JAV08976.1 AAZX01001625 LBMM01000752 KMQ97569.1 KQ981906 KYN33299.1 NNAY01005274 OXU16800.1 GDHC01005538 JAQ13091.1 GECU01006538 JAT01169.1 GL771210 EFZ09794.1 GL887888 EGI69731.1 GBHO01000786 JAG42818.1 FX985558 BBF97892.1 GBRD01006016 JAG59805.1 ACPB03001467 GBBI01002326 JAC16386.1 GBYB01004168 GBYB01004169 GBYB01009716 JAG73935.1 JAG73936.1 JAG79483.1 KQ977600 KYN01518.1 GECZ01012986 JAS56783.1 GBYB01004170 JAG73937.1 QOIP01000002 RLU26078.1 KQ981153 KYN08854.1 GFTR01007850 JAW08576.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000002320
UP000075920
+ More
UP000095301 UP000091820 UP000069940 UP000249989 UP000008820 UP000075883 UP000075902 UP000030765 UP000075840 UP000076407 UP000075903 UP000075882 UP000075885 UP000007062 UP000092443 UP000075900 UP000075884 UP000076408 UP000069272 UP000092444 UP000037069 UP000092460 UP000075881 UP000075880 UP000092445 UP000095300 UP000075901 UP000078200 UP000075886 UP000000673 UP000183832 UP000009192 UP000192221 UP000008792 UP000192223 UP000008711 UP000092553 UP000000304 UP000001292 UP000000803 UP000002282 UP000092461 UP000268350 UP000007801 UP000007266 UP000007798 UP000001070 UP000027135 UP000001819 UP000005204 UP000235965 UP000037510 UP000000311 UP000002358 UP000036403 UP000078541 UP000215335 UP000007755 UP000015103 UP000078542 UP000279307 UP000078492
UP000095301 UP000091820 UP000069940 UP000249989 UP000008820 UP000075883 UP000075902 UP000030765 UP000075840 UP000076407 UP000075903 UP000075882 UP000075885 UP000007062 UP000092443 UP000075900 UP000075884 UP000076408 UP000069272 UP000092444 UP000037069 UP000092460 UP000075881 UP000075880 UP000092445 UP000095300 UP000075901 UP000078200 UP000075886 UP000000673 UP000183832 UP000009192 UP000192221 UP000008792 UP000192223 UP000008711 UP000092553 UP000000304 UP000001292 UP000000803 UP000002282 UP000092461 UP000268350 UP000007801 UP000007266 UP000007798 UP000001070 UP000027135 UP000001819 UP000005204 UP000235965 UP000037510 UP000000311 UP000002358 UP000036403 UP000078541 UP000215335 UP000007755 UP000015103 UP000078542 UP000279307 UP000078492
Pfam
Interpro
IPR005517
Transl_elong_EFG/EF2_IV
+ More
IPR041095 EFG_II
IPR035649 EFG_V
IPR009000 Transl_B-barrel_sf
IPR004161 EFTu-like_2
IPR027417 P-loop_NTPase
IPR020568 Ribosomal_S5_D2-typ_fold
IPR035647 EFG_III/V
IPR031157 G_TR_CS
IPR000640 EFG_V-like
IPR005225 Small_GTP-bd_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR000795 TF_GTP-bd_dom
IPR009022 EFG_III
IPR004540 Transl_elong_EFG/EF2
IPR007258 Vps52
IPR017441 Protein_kinase_ATP_BS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR041095 EFG_II
IPR035649 EFG_V
IPR009000 Transl_B-barrel_sf
IPR004161 EFTu-like_2
IPR027417 P-loop_NTPase
IPR020568 Ribosomal_S5_D2-typ_fold
IPR035647 EFG_III/V
IPR031157 G_TR_CS
IPR000640 EFG_V-like
IPR005225 Small_GTP-bd_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR000795 TF_GTP-bd_dom
IPR009022 EFG_III
IPR004540 Transl_elong_EFG/EF2
IPR007258 Vps52
IPR017441 Protein_kinase_ATP_BS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4JK07
A0A194R253
A0A194PZ19
A0A212F564
A0A1Q3FEM1
B0WGM1
+ More
A0A336MWZ0 A0A182WGI1 A0A336KYM9 A0A1I8MS52 A0A1A9WUF6 A0A182GGX6 Q16S14 A0A023EVY5 A0A182M287 A0A182TUU2 A0A084WU39 A0A182HRY1 A0A182XN79 A0A182VJA8 A0A182KU31 A0A182P4Q7 Q7Q1K8 A0A2K7P6C4 A0A1A9YDN1 A0A182R3S7 A0A182NBQ5 A0A182Y0M7 A0A182F0X9 A0A1B0G1L1 A0A2H1X234 A0A0L0BS00 A0A1B0BKX1 A0A182K0G7 A0A182IX89 A0A1A9Z620 A0A1I8Q4C7 A0A182SQM6 A0A1A9V1H1 A0A182QYP8 W5JI53 A0A1J1I174 A0A2M4ADT9 A0A2M4BF04 T1DQ02 A0A2M4BF99 B4KKD5 A0A2M3Z4A2 A0A2M4BF03 A0A1W4UYC7 B4LS49 A0A1W4X6N8 B3N6A5 A0A0J9QX85 A0A034VEY2 A0A0M4EEV7 B4Q5D5 B4HY41 A0A0K8W112 U5EXB5 Q9VM33 B4NZM7 A0A0A1WI02 W8CD33 A0A1B0CWQ0 A0A3B0JMF7 B3MK91 D6WJJ3 B4MZW9 B4JQM7 A0A067QYF5 Q29N77 H9J9F1 A0A2J7RPP8 A0A1B6EF77 A0A0T6B5D7 A0A0L7L7V4 A0A1Y1M2G3 E2A0M4 A0A1L8DR80 K7JA58 A0A0J7L508 A0A195EZJ3 A0A232EEL7 A0A146LZS1 A0A1B6JPJ1 E9JBK2 F4W861 A0A0A9ZGU0 A0A348G619 A0A0K8T3P1 T1HPG2 A0A023F4E6 A0A0C9RR10 A0A195CLD9 A0A1B6G3A0 A0A0C9R809 A0A3L8E232 A0A195D8V1 A0A224X899
A0A336MWZ0 A0A182WGI1 A0A336KYM9 A0A1I8MS52 A0A1A9WUF6 A0A182GGX6 Q16S14 A0A023EVY5 A0A182M287 A0A182TUU2 A0A084WU39 A0A182HRY1 A0A182XN79 A0A182VJA8 A0A182KU31 A0A182P4Q7 Q7Q1K8 A0A2K7P6C4 A0A1A9YDN1 A0A182R3S7 A0A182NBQ5 A0A182Y0M7 A0A182F0X9 A0A1B0G1L1 A0A2H1X234 A0A0L0BS00 A0A1B0BKX1 A0A182K0G7 A0A182IX89 A0A1A9Z620 A0A1I8Q4C7 A0A182SQM6 A0A1A9V1H1 A0A182QYP8 W5JI53 A0A1J1I174 A0A2M4ADT9 A0A2M4BF04 T1DQ02 A0A2M4BF99 B4KKD5 A0A2M3Z4A2 A0A2M4BF03 A0A1W4UYC7 B4LS49 A0A1W4X6N8 B3N6A5 A0A0J9QX85 A0A034VEY2 A0A0M4EEV7 B4Q5D5 B4HY41 A0A0K8W112 U5EXB5 Q9VM33 B4NZM7 A0A0A1WI02 W8CD33 A0A1B0CWQ0 A0A3B0JMF7 B3MK91 D6WJJ3 B4MZW9 B4JQM7 A0A067QYF5 Q29N77 H9J9F1 A0A2J7RPP8 A0A1B6EF77 A0A0T6B5D7 A0A0L7L7V4 A0A1Y1M2G3 E2A0M4 A0A1L8DR80 K7JA58 A0A0J7L508 A0A195EZJ3 A0A232EEL7 A0A146LZS1 A0A1B6JPJ1 E9JBK2 F4W861 A0A0A9ZGU0 A0A348G619 A0A0K8T3P1 T1HPG2 A0A023F4E6 A0A0C9RR10 A0A195CLD9 A0A1B6G3A0 A0A0C9R809 A0A3L8E232 A0A195D8V1 A0A224X899
PDB
4W29
E-value=2.49919e-158,
Score=1436
Ontologies
GO
Topology
Subcellular location
Mitochondrion
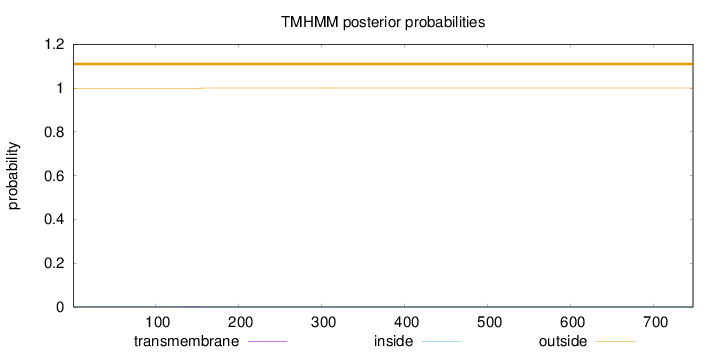
Length:
747
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0410700000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00199
outside
1 - 747
Population Genetic Test Statistics
Pi
185.718917
Theta
158.597291
Tajima's D
0.807044
CLR
0.532606
CSRT
0.609669516524174
Interpretation
Uncertain
