Pre Gene Modal
BGIBMGA006143
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101743906_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.599
Sequence
CDS
ATGCAATTCGAATATGATCCAGAAAAACTCATAGAAGAAGTGGAGAAACGCCCCGGAATTTGGGATTACGATCATGCTGACTATAAAATAAGAAACATACGTTACAAATTGTGGAACGACATTGTAAAAGAACTGATGGAATGCAACGTCAAATTAACAAAAATGGAAATGCGCGAACTCGAAATTCAACTACAAAAGAAATGGAAGAGCATCAAGGATTGTTTTCACAAATACTTGTCGAATCCAAACAAGGGAAGAAGGCCATACATTTACGCAAAAAAATTACAGTTCCTCTTAAAGAACCAAGAGCTGAGGGAAGTAAAAAAGCGAAACAGTGAAAACGCTTCAAGCGAATCGGAAGAAGATACAAAAAGTAAAGTATGGAGACCTAGAAAGAAAATTAAAGTTATCAAATACGAAGACCGTACGTCGGATGAGGATGTAGTCGAAATTGGTGAAAATGATCCACAAGACCAGAGAGACGATTCCAATGACGGAACCGAAGCAAATTCAATGAGGACAGATTCTAAAACAAACAAACCAACGACAGAGGAATTTGCATTCGCGAACGTGGACGTGCCGAAATACGATAACGATGATTCAGACAAAATGTTTCTAATGTCCTTACTACCACATTTAAAATCGGTACGAGAAGAACTAAAGATCAACGTTAAAATGGAACTTATGCAAGTTTTATGTAACGCCAATTATTCAGCAGCCGCTGTTAATAAATTAATCTAA
Protein
MQFEYDPEKLIEEVEKRPGIWDYDHADYKIRNIRYKLWNDIVKELMECNVKLTKMEMRELEIQLQKKWKSIKDCFHKYLSNPNKGRRPYIYAKKLQFLLKNQELREVKKRNSENASSESEEDTKSKVWRPRKKIKVIKYEDRTSDEDVVEIGENDPQDQRDDSNDGTEANSMRTDSKTNKPTTEEFAFANVDVPKYDNDDSDKMFLMSLLPHLKSVREELKINVKMELMQVLCNANYSAAAVNKLI
Summary
Uniprot
A0A2A4JID6
A0A2H1X0E4
A0A0L7KQU0
A0A212FPD0
A0A194Q0Q7
A0A194R3P9
+ More
A0A212EWI2 A0A195BR61 A0A195F3Q1 A0A151XC39 F4WMC5 A0A158P282 A0A195DC77 A0A194QLR2 A0A0J7L3L1 E2AWC0 A0A151IFB9 A0A026WD33 A0A2A3ERT0 A0A088AEC2 A0A194Q210 A0A0N0PC80 A0A154P6K1 A0A0N0BFW2 A0A2S2Q978 A0A2H1WG94 A0A0A9XAN0 A0A0A9X5S0 A0A146KXI5 Q294I7 B4GL21 D0QWN7 A0A2H1WP32 K7IX93 A0A0A9WF21 A0A067RA73 A0A1B6HE62 S4P5P6 A0A3B0JSN8 A0A1B6LQR7 A0A2S2P7V0 A0A1L8FUL7 A0A0N1IJS2 A0A2J7PBC8 T1HA02 A0A0N5D5I8 A0A0P4VZP6 A0A0N0PCI2 A0A2S2NEL4 A0A067R379 A0A1E1WAP8 B4KDM4 A0A1B6JUW1 H9JU56 B4LWV6 A0A0A9YEX1 A0A1E1WBJ4 A0A146LAX9 A0A2P8XKC1 A0A2W1BQ58 A0A194PPX0 A0A0L7QNK6
A0A212EWI2 A0A195BR61 A0A195F3Q1 A0A151XC39 F4WMC5 A0A158P282 A0A195DC77 A0A194QLR2 A0A0J7L3L1 E2AWC0 A0A151IFB9 A0A026WD33 A0A2A3ERT0 A0A088AEC2 A0A194Q210 A0A0N0PC80 A0A154P6K1 A0A0N0BFW2 A0A2S2Q978 A0A2H1WG94 A0A0A9XAN0 A0A0A9X5S0 A0A146KXI5 Q294I7 B4GL21 D0QWN7 A0A2H1WP32 K7IX93 A0A0A9WF21 A0A067RA73 A0A1B6HE62 S4P5P6 A0A3B0JSN8 A0A1B6LQR7 A0A2S2P7V0 A0A1L8FUL7 A0A0N1IJS2 A0A2J7PBC8 T1HA02 A0A0N5D5I8 A0A0P4VZP6 A0A0N0PCI2 A0A2S2NEL4 A0A067R379 A0A1E1WAP8 B4KDM4 A0A1B6JUW1 H9JU56 B4LWV6 A0A0A9YEX1 A0A1E1WBJ4 A0A146LAX9 A0A2P8XKC1 A0A2W1BQ58 A0A194PPX0 A0A0L7QNK6
Pubmed
EMBL
NWSH01001317
PCG71731.1
ODYU01012021
SOQ58104.1
JTDY01007070
KOB65480.1
+ More
AGBW02003328 OWR55560.1 KQ459584 KPI98574.1 KQ460855 KPJ11865.1 AGBW02012010 OWR45804.1 KQ976418 KYM89451.1 KQ981855 KYN34807.1 KQ982315 KYQ57858.1 GL888217 EGI64769.1 ADTU01001164 KQ980989 KYN10515.1 KQ461196 KPJ06467.1 LBMM01000937 KMQ97188.1 GL443280 EFN62280.1 KQ977799 KYM99596.1 KK107261 QOIP01000012 EZA53972.1 RLU16330.1 KZ288192 PBC34400.1 KQ459582 KPI99034.1 KQ460651 KPJ13044.1 KQ434827 KZC07471.1 KQ435794 KOX73672.1 GGMS01005085 MBY74288.1 ODYU01008462 SOQ52093.1 GBHO01029489 JAG14115.1 GBHO01029486 JAG14118.1 GDHC01019159 GDHC01006188 JAP99469.1 JAQ12441.1 CM000070 EAL28977.1 CH479185 EDW38245.1 FJ821033 ACN94736.1 ODYU01010020 SOQ54829.1 AAZX01000580 AAZX01000618 GBHO01040141 GBHO01040140 GBHO01040139 GDHC01012787 GDHC01010302 GDHC01004453 JAG03463.1 JAG03464.1 JAG03465.1 JAQ05842.1 JAQ08327.1 JAQ14176.1 KK852602 KDR20478.1 GECU01034815 JAS72891.1 GAIX01008457 JAA84103.1 OUUW01000008 SPP84023.1 GEBQ01014063 JAT25914.1 GGMR01012912 MBY25531.1 CM004476 OCT75290.1 KQ459825 KPJ19876.1 NEVH01027118 NEVH01016302 NEVH01007440 PNF13628.1 PNF25815.1 PNF35690.1 ACPB03005729 UYYF01004606 VDN05797.1 GDKW01001597 JAI54998.1 KPJ13045.1 GGMR01002773 MBY15392.1 KK852738 KDR17414.1 GDQN01006974 JAT84080.1 CH933806 EDW13858.1 GECU01004715 JAT02992.1 BABH01004109 CH940650 EDW66677.1 GBHO01013966 JAG29638.1 GDQN01006694 JAT84360.1 GDHC01014357 JAQ04272.1 PYGN01001862 PSN32456.1 KZ149935 PZC77192.1 KQ459596 KPI95362.1 KQ414855 KOC60154.1
AGBW02003328 OWR55560.1 KQ459584 KPI98574.1 KQ460855 KPJ11865.1 AGBW02012010 OWR45804.1 KQ976418 KYM89451.1 KQ981855 KYN34807.1 KQ982315 KYQ57858.1 GL888217 EGI64769.1 ADTU01001164 KQ980989 KYN10515.1 KQ461196 KPJ06467.1 LBMM01000937 KMQ97188.1 GL443280 EFN62280.1 KQ977799 KYM99596.1 KK107261 QOIP01000012 EZA53972.1 RLU16330.1 KZ288192 PBC34400.1 KQ459582 KPI99034.1 KQ460651 KPJ13044.1 KQ434827 KZC07471.1 KQ435794 KOX73672.1 GGMS01005085 MBY74288.1 ODYU01008462 SOQ52093.1 GBHO01029489 JAG14115.1 GBHO01029486 JAG14118.1 GDHC01019159 GDHC01006188 JAP99469.1 JAQ12441.1 CM000070 EAL28977.1 CH479185 EDW38245.1 FJ821033 ACN94736.1 ODYU01010020 SOQ54829.1 AAZX01000580 AAZX01000618 GBHO01040141 GBHO01040140 GBHO01040139 GDHC01012787 GDHC01010302 GDHC01004453 JAG03463.1 JAG03464.1 JAG03465.1 JAQ05842.1 JAQ08327.1 JAQ14176.1 KK852602 KDR20478.1 GECU01034815 JAS72891.1 GAIX01008457 JAA84103.1 OUUW01000008 SPP84023.1 GEBQ01014063 JAT25914.1 GGMR01012912 MBY25531.1 CM004476 OCT75290.1 KQ459825 KPJ19876.1 NEVH01027118 NEVH01016302 NEVH01007440 PNF13628.1 PNF25815.1 PNF35690.1 ACPB03005729 UYYF01004606 VDN05797.1 GDKW01001597 JAI54998.1 KPJ13045.1 GGMR01002773 MBY15392.1 KK852738 KDR17414.1 GDQN01006974 JAT84080.1 CH933806 EDW13858.1 GECU01004715 JAT02992.1 BABH01004109 CH940650 EDW66677.1 GBHO01013966 JAG29638.1 GDQN01006694 JAT84360.1 GDHC01014357 JAQ04272.1 PYGN01001862 PSN32456.1 KZ149935 PZC77192.1 KQ459596 KPI95362.1 KQ414855 KOC60154.1
Proteomes
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
UP000078540
+ More
UP000078541 UP000075809 UP000007755 UP000005205 UP000078492 UP000036403 UP000000311 UP000078542 UP000053097 UP000279307 UP000242457 UP000005203 UP000076502 UP000053105 UP000001819 UP000008744 UP000002358 UP000027135 UP000268350 UP000186698 UP000235965 UP000015103 UP000046394 UP000276776 UP000009192 UP000005204 UP000008792 UP000245037 UP000053825
UP000078541 UP000075809 UP000007755 UP000005205 UP000078492 UP000036403 UP000000311 UP000078542 UP000053097 UP000279307 UP000242457 UP000005203 UP000076502 UP000053105 UP000001819 UP000008744 UP000002358 UP000027135 UP000268350 UP000186698 UP000235965 UP000015103 UP000046394 UP000276776 UP000009192 UP000005204 UP000008792 UP000245037 UP000053825
PRIDE
Interpro
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
A0A2A4JID6
A0A2H1X0E4
A0A0L7KQU0
A0A212FPD0
A0A194Q0Q7
A0A194R3P9
+ More
A0A212EWI2 A0A195BR61 A0A195F3Q1 A0A151XC39 F4WMC5 A0A158P282 A0A195DC77 A0A194QLR2 A0A0J7L3L1 E2AWC0 A0A151IFB9 A0A026WD33 A0A2A3ERT0 A0A088AEC2 A0A194Q210 A0A0N0PC80 A0A154P6K1 A0A0N0BFW2 A0A2S2Q978 A0A2H1WG94 A0A0A9XAN0 A0A0A9X5S0 A0A146KXI5 Q294I7 B4GL21 D0QWN7 A0A2H1WP32 K7IX93 A0A0A9WF21 A0A067RA73 A0A1B6HE62 S4P5P6 A0A3B0JSN8 A0A1B6LQR7 A0A2S2P7V0 A0A1L8FUL7 A0A0N1IJS2 A0A2J7PBC8 T1HA02 A0A0N5D5I8 A0A0P4VZP6 A0A0N0PCI2 A0A2S2NEL4 A0A067R379 A0A1E1WAP8 B4KDM4 A0A1B6JUW1 H9JU56 B4LWV6 A0A0A9YEX1 A0A1E1WBJ4 A0A146LAX9 A0A2P8XKC1 A0A2W1BQ58 A0A194PPX0 A0A0L7QNK6
A0A212EWI2 A0A195BR61 A0A195F3Q1 A0A151XC39 F4WMC5 A0A158P282 A0A195DC77 A0A194QLR2 A0A0J7L3L1 E2AWC0 A0A151IFB9 A0A026WD33 A0A2A3ERT0 A0A088AEC2 A0A194Q210 A0A0N0PC80 A0A154P6K1 A0A0N0BFW2 A0A2S2Q978 A0A2H1WG94 A0A0A9XAN0 A0A0A9X5S0 A0A146KXI5 Q294I7 B4GL21 D0QWN7 A0A2H1WP32 K7IX93 A0A0A9WF21 A0A067RA73 A0A1B6HE62 S4P5P6 A0A3B0JSN8 A0A1B6LQR7 A0A2S2P7V0 A0A1L8FUL7 A0A0N1IJS2 A0A2J7PBC8 T1HA02 A0A0N5D5I8 A0A0P4VZP6 A0A0N0PCI2 A0A2S2NEL4 A0A067R379 A0A1E1WAP8 B4KDM4 A0A1B6JUW1 H9JU56 B4LWV6 A0A0A9YEX1 A0A1E1WBJ4 A0A146LAX9 A0A2P8XKC1 A0A2W1BQ58 A0A194PPX0 A0A0L7QNK6
Ontologies
GO
PANTHER
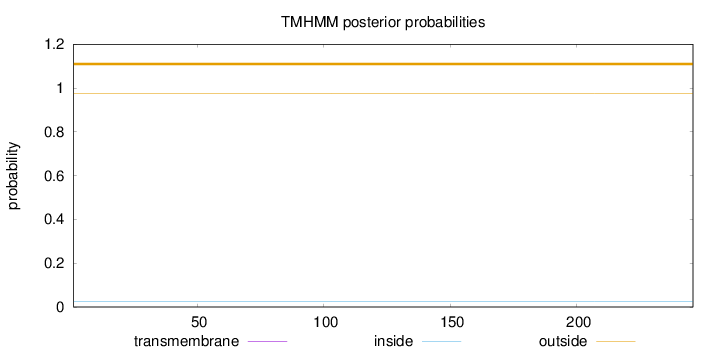
Topology
Subcellular location
Nucleus
Length:
246
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00075
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02565
outside
1 - 246
Population Genetic Test Statistics
Pi
324.538462
Theta
132.554761
Tajima's D
3.611961
CLR
0.253637
CSRT
0.995050247487626
Interpretation
Uncertain
