Gene
KWMTBOMO01756
Pre Gene Modal
BGIBMGA006009
Annotation
PREDICTED:_alsin_[Bombyx_mori]
Full name
Alsin
Alternative Name
Amyotrophic lateral sclerosis 2 protein homolog
Location in the cell
Cytoplasmic Reliability : 3.148
Sequence
CDS
ATGCCCCAAATATCACTAGATTCTAATAGTATTAAGAAGATACCGTTCTTTGAAGGAAGAACTGTATACAGTGCCTCCATTGGTCAAAACTTTGCTGCTGTGATAGCAAGGAAAAATGTTAAACAGTCTGATAACTGCGATACAGACAGTGAAAATGATGAAGAAGAAGTGTTCGCAACTAATTGTTCACAGTGCCAGACTATCATAGGCCTTGCATCTCCTGCATCTGTGCCGTCCTTTTCTGAAACCTGTCCATTAGGTGTTCATATAAGTAAATCAAGTGAAGACTCAAGTAGTACTACTGCTCCACCTACAGATTTTCATATTGATGCTTACAGTTATACTGAAGATTCTAGTCCCTCTTCGGAGGAATCAAAAATAGCACAAAACATTATACAAACTACTGAAACATTAACTGAAAGAAAATGTGATGAATTAATTAATACAGAAAATGGTGATGGCTCAGATAAAGATAGTGATACAACTGAAGCCGATGCAGAAAAGATTAATAATATATTCATGAATGCTGATGCGGCAAGACAGTTTTTAAGTCGACAATTATCATGGGTGTCAGCTGGTGAAGATTATTTAGTGGAATATACAGAAAAACCTACAAGAATCATAAAAGAGAATGTTTCAAATCTCACTAATTTTGTCTATGAAGGTGTTAAAACTGTAGGTGATAAAGTTGCAACACTTTCGAGGCATGTCAGTGGTAGTAGCGAAAACAATGAGACAGTAGAACAACTATCAAGAAATCAGAACGAAGAAACCAAGTCCCTCATGTACAGCTGCACTTCAAACTGTAATGTAGATGATCTGCAGTTTTCCACCAGCACTATTAGTAGTGAAAAAGAGTTTGAAGAAGCCAGAAAAAAAGAGACTATTAAGAACATGTCTAAACTAGGAAGGAATATTTTAGCTACAGAATTATGGACTTGGGGTGACATAAGTTATGGACAGTTAGGAATTGGTGACACAGTCAAAAGGGTTAAACCAATGATTGTCATGAGTTTATCGGGGTTTGGGCTACAAAAAGTTTCTTGTGGGAGTTTGCATTGTTCTACGCTCACGCTCGATGGACGAATGTTTGCTTGGGGATACAATCATTACCATCAATTAAGCTTCGGTTCAAGAGAAGATAAAAGTGGACCACAACAATTTACAGAAAACTCAGAGAGCGATAGAATTAGGGATGTCATTGCTGCTGATCATCATACTCTAGTCTACACGCACAATGAGAATATATTATACATGGGCAAACACACAGATGGTTTCACAGAATCTGTTGTGAATTTAAATCCTAGTTTGTTAGAGGAAGATAGTGAGAAATCTCCAAGCATGGTGACTATAAAAATTACAGAGCCACAACACCATACAAGTAAACCCCAAAATCTGTGTTATCACTGGAGAATCTTATCTTCAGGTAATATTAGTTATTGCTCAGTAGAAAACTCAAAGAAAAATCCACAGTTCCAAGACTTATCTATTGCTCAAAGATATCTAGAAGAAATGTTACTGATTTATCAAAATCTTATTAAGGTTTTTTTGAAGAAAAGTAAAGCAATTACTGCACATTCCAATGTTTATGAAACAGTATGTGATATTTTTGGTGACATATTGAATGTCACAGCTTTAAATGTATTATCAGTTTGGCAGTATGCAGAAAAACATATAAATGAATGTGATATATCATTGATTAAAAATTTAGAAGAGTTTATTTTTTTGTATAGAAAGTATTATGTAGCTGTAAGCAATTTAATTGTTATTGGTGGTTTCACCCAAATCAATAGTGTTATTGATGTCCCAACGAGTATTTATAATTTATTTAGTGATCAACTTCCTTCGAACAAACAGAAAAATAGTAAAAAGACATCAGAAGCCATAATAGGTCTTGCCTTTGTGCAACCATTAACAAGATTAAGTTCATACAAATGTATAACTCAATCTATATTGAGACATAAAACCAAGAGAAAAAAAGGCGAAATTGTATTAAAGATTGAAGAAAGATTAAATAAAGTTGTTTCATCATTTGATTCATTAATAGAAGAGCAAGAAAAAAAACGTAAAGAAGCGGAAGTTACGAAATTATTTTGGGAAACAATTGGTAAAAGTTTGGAGCAATATAGAGCTCCAGAACGACGATTAATAAGAGAATCAAAATCTCGACCTATATATCTAGTCAATCCTGGAAGATTTTCATCACATTGGTTCATTTTATTTAATGACATATTTGTTCATGTTAGTGGTAGCTCACCTAGTGTGCACCCCCTTCAAACCCTGTGGGTCGAGTCTGTCATACATTCTGATCCATCACAATACATAATCCAACTTACTACTCCAGAAGAAGTTATATCAGTGTCAACAGTTACCGAGCAAGAGAAAACTGAATGGATACAAGCTCTGAATAATTCCATAAGAATAATTTTGAACAAAGAATCTGTATTTAAACTACCCTCAATAAGATCAGCAAGTTATACATTCTCAAGTAAAAGCACATTTTTTAAAGATTCGAAATACATTGGTAGATGGTTAGATGGTAAAGTACATGGAAACGGTAAAGTAGAATGGTCAGATGGCAAGCAGTACATTGGACAATTTCAACTAAATACTCTATGTGGCCATGGAAAAATGGAAATACCTGGTGTAGGGATTTATGAAGGCCAGTGGAAAGATAACTTACAGAATGGCTACGGTAAAATGAAGTATTGCTCGGGTGACATATATGAGGGATATTTCAAAGACGGTCAGCCTCATGGACATGGTATTAAAAAGCAAGGAGATTTCACATCTTCATCTGCAACTATTTACACTGGAGAATGGGTTAACGGGGCAAGACAAGGCTACGGTGTCATGGACGACATTGGAAAAGGAGAAAAATATTTAGGAAATTGGAGTGACAACAAGAAACACGGATGTGGGTTGATCGTGACTTTGGACGGAATATATTACGAAGGTTTATTTACGCAAGATGTGTTGACGGGTCATGGGGTTATGGTGTTTGAAGACGGTACACATTACGAAGGCGAATTCAGATCTGCTGGTGTATTTTCTGGTAAAGGAGTTCTCACATTTTCTAGTGGGGATAAGATAGAAGGTTCATTGAGCGGGGCTTGGACTGAGGGAGTTAAAATATCGAATGCCACCATGCATATGAACGTATCGAATCCTGTGTTGCCAGCTAATCCCAAACCTAATTCATTCGGAAAATTAAGCGTAGCACCAAACCAGAAATGGAACGCTCTATTCCGCCAGTGCTTCCAATGCTTGGGTATATCAGACGCGGTGCTAACACCCACCGGTAATCATTTTGCCCACGGACCATCGGGTCCAACGGTGGACAACGTGAAGATATGGCAAAACGTTGCGGTAGCGATATCCTCTTCCCATAAAGAGGGAATGAAAACGAGCCTTAAAAGGAATAAAAGCACTGATCTTGAAAAATCTGTGGATTGTTTGGAAGTCATACCAAAGTTTGGACAAGATACTTTGAATTTAGACGAATATACGGAATTGAAGCAGTATTTGTACAATGCTTGCGAGTCCTCTCACCATCCGCTGGGGCAGCTGGTTCTGGGCTTGACGAACGCATTCAACACGACGTACGGAGGCGTCCGTGTACATCCGCTACTTCTCAGTCATGCGGTCAAAGAACTCAAAAGTATAACCGCGAGGCTCTACCAAGTGGTGAGGCTTCTGTTCCCAGCCCTACCCTCAGAGAATGCCGATCTAGTCTTGCCGTATAAGACCCAAGAGAACGACGATGAAGTTGAAAACGATAGCAATCCTATTCACGGTGAAGTGGTATCGGCAGCTGCATTGCTTCAGCCAACGTTGTTGCCTCGTGTGCACCCGGCTCTATTCGTGCTGTATGCGTTACACAACAAGAGAGAAGACGACATGTACTGGAGGAGGCTGCTGAAATGGAACAAACAGCCGGATATCACTCTGATGGCCTTCCTCGGGATCGATCAGAAATTCTGGGTGAGTTACTCGTCGCAAAACAATCAGTCACCTCCTTACTCTCCGATGAAGGATCAACACTTTCAAGAGGCAGTAGAAACGCTTCAGCAATTGAAGACCACTTTCTCGCCGATCGAGAAACTCCTCGTCATAAGGAATACATTTCAGAAGATGACCAGCGCCGTCCAACAAGAATTAGGTCAAAACTACCTATGGACCATGGACGAGCTGTTCCCGGTCTTCCATTTCGTGGTGGTCCGCTCCAGGATCCTGCAGCTCGGCTCCGAAATACATTTCGTTGAGGACTTCCTCGAGCCGGGCATGCAGCATGGGGAACTCGGCCTTATGTTCACCACTTTAAAGGCCTGCTACTTCCAGATACTTCAAGAGAAGATGTCGATGAATTAG
Protein
MPQISLDSNSIKKIPFFEGRTVYSASIGQNFAAVIARKNVKQSDNCDTDSENDEEEVFATNCSQCQTIIGLASPASVPSFSETCPLGVHISKSSEDSSSTTAPPTDFHIDAYSYTEDSSPSSEESKIAQNIIQTTETLTERKCDELINTENGDGSDKDSDTTEADAEKINNIFMNADAARQFLSRQLSWVSAGEDYLVEYTEKPTRIIKENVSNLTNFVYEGVKTVGDKVATLSRHVSGSSENNETVEQLSRNQNEETKSLMYSCTSNCNVDDLQFSTSTISSEKEFEEARKKETIKNMSKLGRNILATELWTWGDISYGQLGIGDTVKRVKPMIVMSLSGFGLQKVSCGSLHCSTLTLDGRMFAWGYNHYHQLSFGSREDKSGPQQFTENSESDRIRDVIAADHHTLVYTHNENILYMGKHTDGFTESVVNLNPSLLEEDSEKSPSMVTIKITEPQHHTSKPQNLCYHWRILSSGNISYCSVENSKKNPQFQDLSIAQRYLEEMLLIYQNLIKVFLKKSKAITAHSNVYETVCDIFGDILNVTALNVLSVWQYAEKHINECDISLIKNLEEFIFLYRKYYVAVSNLIVIGGFTQINSVIDVPTSIYNLFSDQLPSNKQKNSKKTSEAIIGLAFVQPLTRLSSYKCITQSILRHKTKRKKGEIVLKIEERLNKVVSSFDSLIEEQEKKRKEAEVTKLFWETIGKSLEQYRAPERRLIRESKSRPIYLVNPGRFSSHWFILFNDIFVHVSGSSPSVHPLQTLWVESVIHSDPSQYIIQLTTPEEVISVSTVTEQEKTEWIQALNNSIRIILNKESVFKLPSIRSASYTFSSKSTFFKDSKYIGRWLDGKVHGNGKVEWSDGKQYIGQFQLNTLCGHGKMEIPGVGIYEGQWKDNLQNGYGKMKYCSGDIYEGYFKDGQPHGHGIKKQGDFTSSSATIYTGEWVNGARQGYGVMDDIGKGEKYLGNWSDNKKHGCGLIVTLDGIYYEGLFTQDVLTGHGVMVFEDGTHYEGEFRSAGVFSGKGVLTFSSGDKIEGSLSGAWTEGVKISNATMHMNVSNPVLPANPKPNSFGKLSVAPNQKWNALFRQCFQCLGISDAVLTPTGNHFAHGPSGPTVDNVKIWQNVAVAISSSHKEGMKTSLKRNKSTDLEKSVDCLEVIPKFGQDTLNLDEYTELKQYLYNACESSHHPLGQLVLGLTNAFNTTYGGVRVHPLLLSHAVKELKSITARLYQVVRLLFPALPSENADLVLPYKTQENDDEVENDSNPIHGEVVSAAALLQPTLLPRVHPALFVLYALHNKREDDMYWRRLLKWNKQPDITLMAFLGIDQKFWVSYSSQNNQSPPYSPMKDQHFQEAVETLQQLKTTFSPIEKLLVIRNTFQKMTSAVQQELGQNYLWTMDELFPVFHFVVVRSRILQLGSEIHFVEDFLEPGMQHGELGLMFTTLKACYFQILQEKMSMN
Summary
Description
May act as a GTPase regulator. Controls survival and growth of spinal motoneurons.
May act as a GTPase regulator (By similarity). Controls survival and growth of spinal motoneurons.
May act as a GTPase regulator (By similarity). Controls survival and growth of spinal motoneurons.
Subunit
Forms a heteromeric complex with ALS2CL. Interacts with ALS2CL (By similarity).
Keywords
Acetylation
Alternative splicing
Complete proteome
Guanine-nucleotide releasing factor
Phosphoprotein
Reference proteome
Repeat
Feature
chain Alsin
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J917
A0A2A4IY45
A0A2H1V1E2
A0A194PZ10
A0A194R2W2
A0A0L7LEA1
+ More
E2C623 A0A2A3E6X6 A0A0M8ZRB8 A0A088AJN0 A0A0J7KIQ8 A0A151JZ26 A0A0L7R100 E2AZ33 A0A195BL05 A0A195C4S6 F4WKS2 A0A2J7RB13 A0A026WMF1 A0A151JM59 A0A067RC20 A0A154PSR8 A0A0C9PNX5 A0A232EYP7 A0A1Y1M7V7 D6WJR6 A0A1B6C7L6 A0A1B6BY78 A0A310SV45 A0A023EXX4 T1HUQ5 A0A146M3M8 A0A146LD36 U4U904 N6TET4 N6T7S3 A0A0A9Y1N6 A0A0A9Y655 T1J5J3 A0A1E1X3Y6 A0A1E1XL39 A0A131XRF8 A0A162NJ55 A0A0P5RAZ1 A0A0P5FPW2 A0A2R2MTE0 A0A0P6CN23 A0A210PZY2 A0A2T7PW61 A0A3B4ECD7 A0A3B1IG64 A0A3B4ECG7 Q920R0 H2L2T4 A0A3B5A1S2 A0A146ZQE8 A0A3P9KVA0 A0A3B1JU77 A0A250Y7U7 A0A2D0S6E2 A0A2D0S4V0 A0A1U7QBY6 W5MW38 G1T3G9 W5KC30 A0A1A7WPQ9 A0A3B3UA36 A0A3P9CLZ1 H9G4L6 A0A2I4BGQ4 A0A1A8ECJ3 A0A1A8HZ63 P0C5Y8 W5MW31 A0A1S3MV00 A0A087XQA5 A0A1S3MUT1 A0A146ZR45 A0A3B3VHX5 A0A1A7YZX6 A0A1A8V695 A0A2K6BYF3 A0A2K6A9A2 A0A096MS36 H9FXQ9 A0A2Y9SSC3 G7N8N5 G7PL71 A0A2K6MGE7 A0A2K5M3Z4 A0A2K5REB1 A0A2J8N5A8 A0A2J8WUW9
E2C623 A0A2A3E6X6 A0A0M8ZRB8 A0A088AJN0 A0A0J7KIQ8 A0A151JZ26 A0A0L7R100 E2AZ33 A0A195BL05 A0A195C4S6 F4WKS2 A0A2J7RB13 A0A026WMF1 A0A151JM59 A0A067RC20 A0A154PSR8 A0A0C9PNX5 A0A232EYP7 A0A1Y1M7V7 D6WJR6 A0A1B6C7L6 A0A1B6BY78 A0A310SV45 A0A023EXX4 T1HUQ5 A0A146M3M8 A0A146LD36 U4U904 N6TET4 N6T7S3 A0A0A9Y1N6 A0A0A9Y655 T1J5J3 A0A1E1X3Y6 A0A1E1XL39 A0A131XRF8 A0A162NJ55 A0A0P5RAZ1 A0A0P5FPW2 A0A2R2MTE0 A0A0P6CN23 A0A210PZY2 A0A2T7PW61 A0A3B4ECD7 A0A3B1IG64 A0A3B4ECG7 Q920R0 H2L2T4 A0A3B5A1S2 A0A146ZQE8 A0A3P9KVA0 A0A3B1JU77 A0A250Y7U7 A0A2D0S6E2 A0A2D0S4V0 A0A1U7QBY6 W5MW38 G1T3G9 W5KC30 A0A1A7WPQ9 A0A3B3UA36 A0A3P9CLZ1 H9G4L6 A0A2I4BGQ4 A0A1A8ECJ3 A0A1A8HZ63 P0C5Y8 W5MW31 A0A1S3MV00 A0A087XQA5 A0A1S3MUT1 A0A146ZR45 A0A3B3VHX5 A0A1A7YZX6 A0A1A8V695 A0A2K6BYF3 A0A2K6A9A2 A0A096MS36 H9FXQ9 A0A2Y9SSC3 G7N8N5 G7PL71 A0A2K6MGE7 A0A2K5M3Z4 A0A2K5REB1 A0A2J8N5A8 A0A2J8WUW9
Pubmed
19121390
26354079
26227816
20798317
21719571
24508170
+ More
30249741 24845553 28648823 28004739 18362917 19820115 25474469 26823975 23537049 25401762 28503490 29209593 29652888 28812685 25329095 11586298 19468303 15489334 16141072 15345747 17242355 21183079 23806337 17554307 28087693 21993624 25186727 21881562 15057822 16802292 15686953 22673903 25319552 22002653
30249741 24845553 28648823 28004739 18362917 19820115 25474469 26823975 23537049 25401762 28503490 29209593 29652888 28812685 25329095 11586298 19468303 15489334 16141072 15345747 17242355 21183079 23806337 17554307 28087693 21993624 25186727 21881562 15057822 16802292 15686953 22673903 25319552 22002653
EMBL
BABH01004646
NWSH01004947
PCG64569.1
ODYU01000050
SOQ34162.1
KQ459584
+ More
KPI98567.1 KQ460855 KPJ11859.1 JTDY01001500 KOB73720.1 GL452895 EFN76628.1 KZ288349 PBC27450.1 KQ435964 KOX67949.1 LBMM01006796 KMQ90323.1 KQ981427 KYN41758.1 KQ414668 KOC64530.1 GL444078 EFN61307.1 KQ976453 KYM85362.1 KQ978317 KYM95163.1 GL888206 EGI65164.1 NEVH01006564 PNF38027.1 KK107167 QOIP01000010 EZA56294.1 RLU17499.1 KQ978949 KYN27375.1 KK852804 KDR16248.1 KQ435178 KZC14951.1 GBYB01001400 GBYB01002833 JAG71167.1 JAG72600.1 NNAY01001596 OXU23475.1 GEZM01040965 JAV80680.1 KQ971343 EFA04485.1 GEDC01027979 JAS09319.1 GEDC01031077 JAS06221.1 KQ760158 OAD61694.1 GBBI01004557 JAC14155.1 ACPB03002623 GDHC01005007 JAQ13622.1 GDHC01013573 JAQ05056.1 KB631904 ERL87101.1 APGK01040756 KB740984 ENN76273.1 ENN76274.1 GBHO01018581 GBHO01018577 JAG25023.1 JAG25027.1 GBHO01018583 GBHO01018579 JAG25021.1 JAG25025.1 JH431865 GFAC01005213 JAT93975.1 GFAA01003510 JAT99924.1 GEFM01005923 GEGO01005350 JAP69873.1 JAR90054.1 LRGB01000568 KZS18031.1 GDIQ01122860 JAL28866.1 GDIP01145376 JAJ78026.1 GDIP01012169 JAM91546.1 NEDP02005321 OWF42064.1 PZQS01000001 PVD37648.1 AB053307 AC153652 CH466548 BC031479 BC046828 AK014320 AAH31479.1 AAH46828.1 BAB69016.1 GCES01017773 JAR68550.1 GFFV01000238 JAV39707.1 AHAT01034650 AHAT01034651 AAGW02042203 AAGW02042204 AAGW02042205 HADW01006119 SBP07519.1 AAWZ02028705 AAWZ02028706 HAEA01014846 SBQ43326.1 HAED01003547 SBQ89530.1 AABR03068212 BK005190 AYCK01006857 GCES01017512 JAR68811.1 HADX01013444 SBP35676.1 HAEJ01014301 SBS54758.1 AHZZ02004391 JU335664 JU335665 JU335666 JU473719 JU473720 AFE79418.1 AFH30523.1 CM001264 EHH21585.1 AQIA01015563 AQIA01015564 AQIA01015565 CM001287 EHH55080.1 NBAG03000236 PNI66951.1 NDHI03003377 PNJ73577.1
KPI98567.1 KQ460855 KPJ11859.1 JTDY01001500 KOB73720.1 GL452895 EFN76628.1 KZ288349 PBC27450.1 KQ435964 KOX67949.1 LBMM01006796 KMQ90323.1 KQ981427 KYN41758.1 KQ414668 KOC64530.1 GL444078 EFN61307.1 KQ976453 KYM85362.1 KQ978317 KYM95163.1 GL888206 EGI65164.1 NEVH01006564 PNF38027.1 KK107167 QOIP01000010 EZA56294.1 RLU17499.1 KQ978949 KYN27375.1 KK852804 KDR16248.1 KQ435178 KZC14951.1 GBYB01001400 GBYB01002833 JAG71167.1 JAG72600.1 NNAY01001596 OXU23475.1 GEZM01040965 JAV80680.1 KQ971343 EFA04485.1 GEDC01027979 JAS09319.1 GEDC01031077 JAS06221.1 KQ760158 OAD61694.1 GBBI01004557 JAC14155.1 ACPB03002623 GDHC01005007 JAQ13622.1 GDHC01013573 JAQ05056.1 KB631904 ERL87101.1 APGK01040756 KB740984 ENN76273.1 ENN76274.1 GBHO01018581 GBHO01018577 JAG25023.1 JAG25027.1 GBHO01018583 GBHO01018579 JAG25021.1 JAG25025.1 JH431865 GFAC01005213 JAT93975.1 GFAA01003510 JAT99924.1 GEFM01005923 GEGO01005350 JAP69873.1 JAR90054.1 LRGB01000568 KZS18031.1 GDIQ01122860 JAL28866.1 GDIP01145376 JAJ78026.1 GDIP01012169 JAM91546.1 NEDP02005321 OWF42064.1 PZQS01000001 PVD37648.1 AB053307 AC153652 CH466548 BC031479 BC046828 AK014320 AAH31479.1 AAH46828.1 BAB69016.1 GCES01017773 JAR68550.1 GFFV01000238 JAV39707.1 AHAT01034650 AHAT01034651 AAGW02042203 AAGW02042204 AAGW02042205 HADW01006119 SBP07519.1 AAWZ02028705 AAWZ02028706 HAEA01014846 SBQ43326.1 HAED01003547 SBQ89530.1 AABR03068212 BK005190 AYCK01006857 GCES01017512 JAR68811.1 HADX01013444 SBP35676.1 HAEJ01014301 SBS54758.1 AHZZ02004391 JU335664 JU335665 JU335666 JU473719 JU473720 AFE79418.1 AFH30523.1 CM001264 EHH21585.1 AQIA01015563 AQIA01015564 AQIA01015565 CM001287 EHH55080.1 NBAG03000236 PNI66951.1 NDHI03003377 PNJ73577.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000008237
+ More
UP000242457 UP000053105 UP000005203 UP000036403 UP000078541 UP000053825 UP000000311 UP000078540 UP000078542 UP000007755 UP000235965 UP000053097 UP000279307 UP000078492 UP000027135 UP000076502 UP000215335 UP000007266 UP000015103 UP000030742 UP000019118 UP000076858 UP000085678 UP000242188 UP000245119 UP000261440 UP000018467 UP000000589 UP000001038 UP000261400 UP000265180 UP000221080 UP000189706 UP000018468 UP000001811 UP000261500 UP000265160 UP000001646 UP000192220 UP000002494 UP000087266 UP000028760 UP000233120 UP000233140 UP000028761 UP000248484 UP000009130 UP000233100 UP000233180 UP000233060 UP000233040
UP000242457 UP000053105 UP000005203 UP000036403 UP000078541 UP000053825 UP000000311 UP000078540 UP000078542 UP000007755 UP000235965 UP000053097 UP000279307 UP000078492 UP000027135 UP000076502 UP000215335 UP000007266 UP000015103 UP000030742 UP000019118 UP000076858 UP000085678 UP000242188 UP000245119 UP000261440 UP000018467 UP000000589 UP000001038 UP000261400 UP000265180 UP000221080 UP000189706 UP000018468 UP000001811 UP000261500 UP000265160 UP000001646 UP000192220 UP000002494 UP000087266 UP000028760 UP000233120 UP000233140 UP000028761 UP000248484 UP000009130 UP000233100 UP000233180 UP000233060 UP000233040
Pfam
Interpro
IPR003123
VPS9
+ More
IPR003409 MORN
IPR037191 VPS9_dom_sf
IPR000408 Reg_chr_condens
IPR011993 PH-like_dom_sf
IPR009091 RCC1/BLIP-II
IPR001849 PH_domain
IPR035899 DBL_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR005636 DTW
IPR000219 DH-domain
IPR036451 CblAdoTrfase-like_sf
IPR016030 CblAdoTrfase-like
IPR029499 PduO-typ
IPR001930 Peptidase_M1
IPR014830 Glycolipid_transfer_prot_dom
IPR034016 M1_APN-typ
IPR024571 ERAP1-like_C_dom
IPR014782 Peptidase_M1_dom
IPR042097 Aminopeptidase_N-like_N
IPR036497 GLTP_sf
IPR003409 MORN
IPR037191 VPS9_dom_sf
IPR000408 Reg_chr_condens
IPR011993 PH-like_dom_sf
IPR009091 RCC1/BLIP-II
IPR001849 PH_domain
IPR035899 DBL_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR005636 DTW
IPR000219 DH-domain
IPR036451 CblAdoTrfase-like_sf
IPR016030 CblAdoTrfase-like
IPR029499 PduO-typ
IPR001930 Peptidase_M1
IPR014830 Glycolipid_transfer_prot_dom
IPR034016 M1_APN-typ
IPR024571 ERAP1-like_C_dom
IPR014782 Peptidase_M1_dom
IPR042097 Aminopeptidase_N-like_N
IPR036497 GLTP_sf
SUPFAM
CDD
ProteinModelPortal
H9J917
A0A2A4IY45
A0A2H1V1E2
A0A194PZ10
A0A194R2W2
A0A0L7LEA1
+ More
E2C623 A0A2A3E6X6 A0A0M8ZRB8 A0A088AJN0 A0A0J7KIQ8 A0A151JZ26 A0A0L7R100 E2AZ33 A0A195BL05 A0A195C4S6 F4WKS2 A0A2J7RB13 A0A026WMF1 A0A151JM59 A0A067RC20 A0A154PSR8 A0A0C9PNX5 A0A232EYP7 A0A1Y1M7V7 D6WJR6 A0A1B6C7L6 A0A1B6BY78 A0A310SV45 A0A023EXX4 T1HUQ5 A0A146M3M8 A0A146LD36 U4U904 N6TET4 N6T7S3 A0A0A9Y1N6 A0A0A9Y655 T1J5J3 A0A1E1X3Y6 A0A1E1XL39 A0A131XRF8 A0A162NJ55 A0A0P5RAZ1 A0A0P5FPW2 A0A2R2MTE0 A0A0P6CN23 A0A210PZY2 A0A2T7PW61 A0A3B4ECD7 A0A3B1IG64 A0A3B4ECG7 Q920R0 H2L2T4 A0A3B5A1S2 A0A146ZQE8 A0A3P9KVA0 A0A3B1JU77 A0A250Y7U7 A0A2D0S6E2 A0A2D0S4V0 A0A1U7QBY6 W5MW38 G1T3G9 W5KC30 A0A1A7WPQ9 A0A3B3UA36 A0A3P9CLZ1 H9G4L6 A0A2I4BGQ4 A0A1A8ECJ3 A0A1A8HZ63 P0C5Y8 W5MW31 A0A1S3MV00 A0A087XQA5 A0A1S3MUT1 A0A146ZR45 A0A3B3VHX5 A0A1A7YZX6 A0A1A8V695 A0A2K6BYF3 A0A2K6A9A2 A0A096MS36 H9FXQ9 A0A2Y9SSC3 G7N8N5 G7PL71 A0A2K6MGE7 A0A2K5M3Z4 A0A2K5REB1 A0A2J8N5A8 A0A2J8WUW9
E2C623 A0A2A3E6X6 A0A0M8ZRB8 A0A088AJN0 A0A0J7KIQ8 A0A151JZ26 A0A0L7R100 E2AZ33 A0A195BL05 A0A195C4S6 F4WKS2 A0A2J7RB13 A0A026WMF1 A0A151JM59 A0A067RC20 A0A154PSR8 A0A0C9PNX5 A0A232EYP7 A0A1Y1M7V7 D6WJR6 A0A1B6C7L6 A0A1B6BY78 A0A310SV45 A0A023EXX4 T1HUQ5 A0A146M3M8 A0A146LD36 U4U904 N6TET4 N6T7S3 A0A0A9Y1N6 A0A0A9Y655 T1J5J3 A0A1E1X3Y6 A0A1E1XL39 A0A131XRF8 A0A162NJ55 A0A0P5RAZ1 A0A0P5FPW2 A0A2R2MTE0 A0A0P6CN23 A0A210PZY2 A0A2T7PW61 A0A3B4ECD7 A0A3B1IG64 A0A3B4ECG7 Q920R0 H2L2T4 A0A3B5A1S2 A0A146ZQE8 A0A3P9KVA0 A0A3B1JU77 A0A250Y7U7 A0A2D0S6E2 A0A2D0S4V0 A0A1U7QBY6 W5MW38 G1T3G9 W5KC30 A0A1A7WPQ9 A0A3B3UA36 A0A3P9CLZ1 H9G4L6 A0A2I4BGQ4 A0A1A8ECJ3 A0A1A8HZ63 P0C5Y8 W5MW31 A0A1S3MV00 A0A087XQA5 A0A1S3MUT1 A0A146ZR45 A0A3B3VHX5 A0A1A7YZX6 A0A1A8V695 A0A2K6BYF3 A0A2K6A9A2 A0A096MS36 H9FXQ9 A0A2Y9SSC3 G7N8N5 G7PL71 A0A2K6MGE7 A0A2K5M3Z4 A0A2K5REB1 A0A2J8N5A8 A0A2J8WUW9
PDB
4DNV
E-value=6.82304e-12,
Score=176
Ontologies
GO
GO:0005089
GO:0035023
GO:0005737
GO:0008237
GO:0120013
GO:0008270
GO:0043005
GO:0016197
GO:0030424
GO:0030425
GO:0030676
GO:0031982
GO:0030426
GO:0045860
GO:0001662
GO:0007409
GO:0014069
GO:0043547
GO:0005813
GO:0051036
GO:0030027
GO:0042803
GO:0043231
GO:0007032
GO:0006979
GO:0016020
GO:0042802
GO:0017137
GO:0035249
GO:0001881
GO:0043539
GO:0005634
GO:0043087
GO:0016601
GO:0007626
GO:0016050
GO:0008219
GO:0043025
GO:0005829
GO:0048365
GO:0048812
GO:0001726
GO:0051260
GO:0005096
GO:0001701
GO:0043197
GO:0007528
GO:0032991
GO:0005769
GO:0008104
GO:0017112
GO:0007041
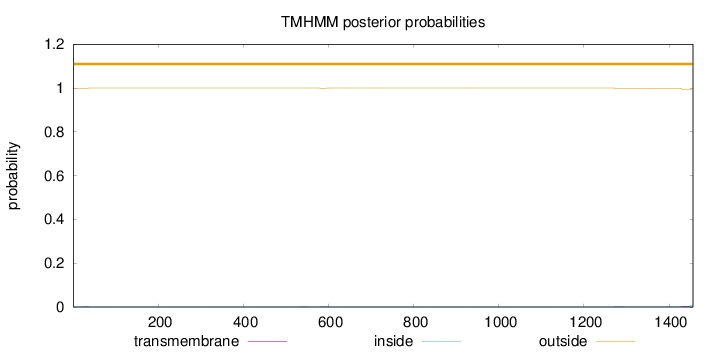
Topology
Length:
1456
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13812
Exp number, first 60 AAs:
0.00415
Total prob of N-in:
0.00042
outside
1 - 1456
Population Genetic Test Statistics
Pi
199.796803
Theta
167.585102
Tajima's D
0.492938
CLR
0.393216
CSRT
0.507524623768812
Interpretation
Uncertain
