Gene
KWMTBOMO01755
Pre Gene Modal
BGIBMGA006140
Annotation
PREDICTED:_carboxy-terminal_domain_RNA_polymerase_II_polypeptide_A_small_phosphatase_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.605 PlasmaMembrane Reliability : 1.078
Sequence
CDS
ATGGACGCATCGTCCATTATAACTCAGGTGTCACGGGATGACGAACAATTAAACAATTATGGACCCGATAGAGGTTCATCCCCTGGTGTGCAGCATAATGAGGGTGACGGCACCCCAGTTTCGGGGTCGACCCCTTTGGGTGGGAAGAAATCTAGCGGGACAGGCGGCTTCTTGCGATCGTTGCTGTGCTGTTGGCGCGGTGGCCGAGGCAAGGGACCTCCCGGAGGCAATGGCACCAATAGTATCGACGGCAGGGCTTCTCCGCCCTTGCTGGTCATGTCTGAACATGCCCCCAGACCTCTGTTACCGCCAGTGAGACACCAGGACATGCATAAAAAGTGTATGGTTATTGATTTGGATGAAACATTAGTGCACAGCTCATTCAAGCCTATAAACAATGCAGATTTCGTGGTGCCTGTAGAAATCGACGGTGCGGTCCATCAAGTTTACGTTCTGAAGAGGCCGCATGTCGACGAGTTCCTGCGGCGGTGTGGCGAGCTTTACGAGTGCGTTTTGTTCACCGCGTCATTGGCGAAGTACGCCGATCCTGTCGCCGATCTACTGGACCGATGGGGCGTATTCCGCGCGCGGCTGTTCCGCGACAGCTGCGTGTTCCACCGCGGCAACTACGTCAAGGACCTGAACAGCCTCGGGCGGGACCTGCGGCGGGTCGTCATCGTCGACAACTCGCCCGCTTCTTACATATTCCACCCGGATAATGCTGTGCCTGTCGCGTCCTGGTTCGACGACATGACAGATTCTGAGCTGCTCGACCTGATCCCGTTCTTCGAGAAGCTGAGCAAAGTGGAGAGCGTGTACACTGTCTTGCGCAACTCGAACCACCCGTACAACCCGACGCAGAACAACTCGCCGAGCACATAG
Protein
MDASSIITQVSRDDEQLNNYGPDRGSSPGVQHNEGDGTPVSGSTPLGGKKSSGTGGFLRSLLCCWRGGRGKGPPGGNGTNSIDGRASPPLLVMSEHAPRPLLPPVRHQDMHKKCMVIDLDETLVHSSFKPINNADFVVPVEIDGAVHQVYVLKRPHVDEFLRRCGELYECVLFTASLAKYADPVADLLDRWGVFRARLFRDSCVFHRGNYVKDLNSLGRDLRRVVIVDNSPASYIFHPDNAVPVASWFDDMTDSELLDLIPFFEKLSKVESVYTVLRNSNHPYNPTQNNSPST
Summary
Uniprot
A0A194Q591
A0A212EVW7
A0A2A4JQV9
A0A194R243
A0A2A4JP74
A0A1E1WIZ3
+ More
A0A1B6CPC4 A0A2P8Z7T5 A0A1W4XCY2 A0A1Y1KLL5 A0A0M8ZX50 A0A1B6JIN2 A0A1B6MJA2 A0A088AD85 E2BAZ1 A0A224XV12 A0A023F553 A0A067R0I1 N6UC94 A0A195EZI6 A0A0N7Z8X6 D6WJR7 A0A158P2R1 A0A0P4VPQ7 A0A026VU91 A0A0C9Q259 A0A0A9X8N7 K7IXM6 E0VJR2 A0A0A9W7P1 A0A0J7KML9 E9IPJ7 A0A336K4Q7 F4W5S0 A0A195DGJ8 A0A3Q0J7Z6 A0A151X2K9 B4LDZ8 A0A0Q9WUW3 B4KYH7 B4J3N7 A0A1L8DGM0 A0A154PEV8 A0A0A1WYU9 A0A3B0K0A8 A0A034W076 W8BEZ4 A0A034W4C9 A0A0A1WLG9 B4H7T2 Q29D94 A0A1W4VMF8 B3NDQ2 B3M9T8 A0A0K8V083 A0A2A3ECH6 B4ITV4 U4UAW4 A0A1Z5LDW7 Q9VUX0 A0A0J9RW12 B4QLS5 A0A1Q3FCD0 V5IFU9 A0A0K8TR68 Q7YU53 M9PFN0 A0A023FJT2 A0A023GK94 G3MP86 J9JLI7 A0A2S2QHR5 A0A131XFW3 A0A131ZDT6 L7M156 A0A2S2PNP6 A0A2R5LJT7 A0A2H8TEC6 A0A293M2H0 A0A1I8JVR3 A0A2M4CPD0 A0A0K8RRI2 A0A1I8MRU9 B4N728 A0A210QY34 E9G5C4 A0A232F0E4 A0A023FYD6 A0A2R7X5F7 A0A1E1X2F2 C3YYP1 A0A224YV46 A0A1B0GCP0 B4HIM4 A0A162NJ95 A0A2C9JQ61
A0A1B6CPC4 A0A2P8Z7T5 A0A1W4XCY2 A0A1Y1KLL5 A0A0M8ZX50 A0A1B6JIN2 A0A1B6MJA2 A0A088AD85 E2BAZ1 A0A224XV12 A0A023F553 A0A067R0I1 N6UC94 A0A195EZI6 A0A0N7Z8X6 D6WJR7 A0A158P2R1 A0A0P4VPQ7 A0A026VU91 A0A0C9Q259 A0A0A9X8N7 K7IXM6 E0VJR2 A0A0A9W7P1 A0A0J7KML9 E9IPJ7 A0A336K4Q7 F4W5S0 A0A195DGJ8 A0A3Q0J7Z6 A0A151X2K9 B4LDZ8 A0A0Q9WUW3 B4KYH7 B4J3N7 A0A1L8DGM0 A0A154PEV8 A0A0A1WYU9 A0A3B0K0A8 A0A034W076 W8BEZ4 A0A034W4C9 A0A0A1WLG9 B4H7T2 Q29D94 A0A1W4VMF8 B3NDQ2 B3M9T8 A0A0K8V083 A0A2A3ECH6 B4ITV4 U4UAW4 A0A1Z5LDW7 Q9VUX0 A0A0J9RW12 B4QLS5 A0A1Q3FCD0 V5IFU9 A0A0K8TR68 Q7YU53 M9PFN0 A0A023FJT2 A0A023GK94 G3MP86 J9JLI7 A0A2S2QHR5 A0A131XFW3 A0A131ZDT6 L7M156 A0A2S2PNP6 A0A2R5LJT7 A0A2H8TEC6 A0A293M2H0 A0A1I8JVR3 A0A2M4CPD0 A0A0K8RRI2 A0A1I8MRU9 B4N728 A0A210QY34 E9G5C4 A0A232F0E4 A0A023FYD6 A0A2R7X5F7 A0A1E1X2F2 C3YYP1 A0A224YV46 A0A1B0GCP0 B4HIM4 A0A162NJ95 A0A2C9JQ61
Pubmed
26354079
22118469
29403074
28004739
20798317
25474469
+ More
24845553 23537049 27129103 18362917 19820115 21347285 24508170 30249741 25401762 26823975 20075255 20566863 21282665 21719571 17994087 25830018 25348373 24495485 15632085 23185243 17550304 28528879 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25765539 26369729 22216098 28049606 26830274 25576852 25315136 28812685 21292972 28648823 28503490 18563158 28797301 15562597
24845553 23537049 27129103 18362917 19820115 21347285 24508170 30249741 25401762 26823975 20075255 20566863 21282665 21719571 17994087 25830018 25348373 24495485 15632085 23185243 17550304 28528879 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25765539 26369729 22216098 28049606 26830274 25576852 25315136 28812685 21292972 28648823 28503490 18563158 28797301 15562597
EMBL
KQ459584
KPI98565.1
AGBW02012119
OWR45611.1
NWSH01000867
PCG73802.1
+ More
KQ460855 KPJ11858.1 PCG73801.1 GDQN01004146 GDQN01002564 JAT86908.1 JAT88490.1 GEDC01022067 JAS15231.1 PYGN01000158 PSN52556.1 GEZM01080349 JAV62204.1 KQ435811 KOX72905.1 GECU01008550 JAS99156.1 GEBQ01004012 JAT35965.1 GL446901 EFN87082.1 GFTR01004655 JAW11771.1 GBBI01002514 JAC16198.1 KK852804 KDR16246.1 APGK01040754 APGK01040755 KB740984 ENN76272.1 KQ981905 KYN33327.1 GDKW01002123 JAI54472.1 KQ971343 EFA03107.1 ADTU01007394 GDKW01002122 JAI54473.1 KK107921 QOIP01000001 EZA47215.1 RLU26385.1 GBYB01008093 JAG77860.1 GBHO01028466 GBRD01015826 GDHC01014324 JAG15138.1 JAG50000.1 JAQ04305.1 AAZX01014366 AAZX01019770 DS235226 EEB13618.1 GBHO01042729 GBRD01015827 GDHC01002898 JAG00875.1 JAG49999.1 JAQ15731.1 LBMM01005397 KMQ91552.1 GL764539 EFZ17505.1 UFQS01000099 UFQT01000099 SSW99497.1 SSX19877.1 GL887695 EGI70396.1 KQ980903 KYN11589.1 KQ982576 KYQ54675.1 CH940647 EDW70041.1 KRF84686.1 CH933809 EDW18788.2 CH916366 EDV97268.1 GFDF01008584 JAV05500.1 KQ434889 KZC10389.1 GBXI01010597 JAD03695.1 OUUW01000002 SPP76798.1 GAKP01009966 JAC48986.1 GAMC01009313 JAB97242.1 GAKP01009967 JAC48985.1 GBXI01014590 JAC99701.1 CH479219 EDW34722.1 CH379070 EAL30520.2 KRT09029.1 CH954178 EDV52185.1 CH902618 EDV40129.1 GDHF01020000 JAI32314.1 KZ288285 PBC29483.1 CH891734 EDW99817.1 KB631904 ERL87100.1 GFJQ02001839 JAW05131.1 AE014296 BT050447 AAF49553.2 ACJ13154.1 CM002912 KMY99893.1 CM000363 EDX10624.1 GFDL01009810 JAV25235.1 GANP01007807 JAB76661.1 GDAI01001183 JAI16420.1 BT009984 AAQ22453.1 AGB94605.1 GBBK01003177 JAC21305.1 GBBM01001975 JAC33443.1 JO843687 AEO35304.1 ABLF02032746 GGMS01008106 GGMS01011690 MBY77309.1 MBY80893.1 GEFH01003980 JAP64601.1 GEDV01000156 JAP88401.1 GACK01006993 JAA58041.1 GGMR01018450 MBY31069.1 GGLE01005613 MBY09739.1 GFXV01000652 MBW12457.1 GFWV01010444 MAA35173.1 GGFL01002999 MBW67177.1 GADI01000137 JAA73671.1 CH964168 EDW80167.1 NEDP02001306 OWF53612.1 GL732532 EFX85656.1 NNAY01001405 OXU24082.1 GBBL01001595 JAC25725.1 KK857164 PTY26901.1 GFAC01005751 JAT93437.1 GG666565 EEN54681.1 GFPF01006556 MAA17702.1 CCAG010002173 CCAG010002174 CCAG010002175 CH480815 EDW41654.1 LRGB01000568 KZS18035.1
KQ460855 KPJ11858.1 PCG73801.1 GDQN01004146 GDQN01002564 JAT86908.1 JAT88490.1 GEDC01022067 JAS15231.1 PYGN01000158 PSN52556.1 GEZM01080349 JAV62204.1 KQ435811 KOX72905.1 GECU01008550 JAS99156.1 GEBQ01004012 JAT35965.1 GL446901 EFN87082.1 GFTR01004655 JAW11771.1 GBBI01002514 JAC16198.1 KK852804 KDR16246.1 APGK01040754 APGK01040755 KB740984 ENN76272.1 KQ981905 KYN33327.1 GDKW01002123 JAI54472.1 KQ971343 EFA03107.1 ADTU01007394 GDKW01002122 JAI54473.1 KK107921 QOIP01000001 EZA47215.1 RLU26385.1 GBYB01008093 JAG77860.1 GBHO01028466 GBRD01015826 GDHC01014324 JAG15138.1 JAG50000.1 JAQ04305.1 AAZX01014366 AAZX01019770 DS235226 EEB13618.1 GBHO01042729 GBRD01015827 GDHC01002898 JAG00875.1 JAG49999.1 JAQ15731.1 LBMM01005397 KMQ91552.1 GL764539 EFZ17505.1 UFQS01000099 UFQT01000099 SSW99497.1 SSX19877.1 GL887695 EGI70396.1 KQ980903 KYN11589.1 KQ982576 KYQ54675.1 CH940647 EDW70041.1 KRF84686.1 CH933809 EDW18788.2 CH916366 EDV97268.1 GFDF01008584 JAV05500.1 KQ434889 KZC10389.1 GBXI01010597 JAD03695.1 OUUW01000002 SPP76798.1 GAKP01009966 JAC48986.1 GAMC01009313 JAB97242.1 GAKP01009967 JAC48985.1 GBXI01014590 JAC99701.1 CH479219 EDW34722.1 CH379070 EAL30520.2 KRT09029.1 CH954178 EDV52185.1 CH902618 EDV40129.1 GDHF01020000 JAI32314.1 KZ288285 PBC29483.1 CH891734 EDW99817.1 KB631904 ERL87100.1 GFJQ02001839 JAW05131.1 AE014296 BT050447 AAF49553.2 ACJ13154.1 CM002912 KMY99893.1 CM000363 EDX10624.1 GFDL01009810 JAV25235.1 GANP01007807 JAB76661.1 GDAI01001183 JAI16420.1 BT009984 AAQ22453.1 AGB94605.1 GBBK01003177 JAC21305.1 GBBM01001975 JAC33443.1 JO843687 AEO35304.1 ABLF02032746 GGMS01008106 GGMS01011690 MBY77309.1 MBY80893.1 GEFH01003980 JAP64601.1 GEDV01000156 JAP88401.1 GACK01006993 JAA58041.1 GGMR01018450 MBY31069.1 GGLE01005613 MBY09739.1 GFXV01000652 MBW12457.1 GFWV01010444 MAA35173.1 GGFL01002999 MBW67177.1 GADI01000137 JAA73671.1 CH964168 EDW80167.1 NEDP02001306 OWF53612.1 GL732532 EFX85656.1 NNAY01001405 OXU24082.1 GBBL01001595 JAC25725.1 KK857164 PTY26901.1 GFAC01005751 JAT93437.1 GG666565 EEN54681.1 GFPF01006556 MAA17702.1 CCAG010002173 CCAG010002174 CCAG010002175 CH480815 EDW41654.1 LRGB01000568 KZS18035.1
Proteomes
UP000053268
UP000007151
UP000218220
UP000053240
UP000245037
UP000192223
+ More
UP000053105 UP000005203 UP000008237 UP000027135 UP000019118 UP000078541 UP000007266 UP000005205 UP000053097 UP000279307 UP000002358 UP000009046 UP000036403 UP000007755 UP000078492 UP000079169 UP000075809 UP000008792 UP000009192 UP000001070 UP000076502 UP000268350 UP000008744 UP000001819 UP000192221 UP000008711 UP000007801 UP000242457 UP000002282 UP000030742 UP000000803 UP000000304 UP000007819 UP000076407 UP000095301 UP000007798 UP000242188 UP000000305 UP000215335 UP000001554 UP000092444 UP000001292 UP000076858 UP000076420
UP000053105 UP000005203 UP000008237 UP000027135 UP000019118 UP000078541 UP000007266 UP000005205 UP000053097 UP000279307 UP000002358 UP000009046 UP000036403 UP000007755 UP000078492 UP000079169 UP000075809 UP000008792 UP000009192 UP000001070 UP000076502 UP000268350 UP000008744 UP000001819 UP000192221 UP000008711 UP000007801 UP000242457 UP000002282 UP000030742 UP000000803 UP000000304 UP000007819 UP000076407 UP000095301 UP000007798 UP000242188 UP000000305 UP000215335 UP000001554 UP000092444 UP000001292 UP000076858 UP000076420
Interpro
Gene 3D
ProteinModelPortal
A0A194Q591
A0A212EVW7
A0A2A4JQV9
A0A194R243
A0A2A4JP74
A0A1E1WIZ3
+ More
A0A1B6CPC4 A0A2P8Z7T5 A0A1W4XCY2 A0A1Y1KLL5 A0A0M8ZX50 A0A1B6JIN2 A0A1B6MJA2 A0A088AD85 E2BAZ1 A0A224XV12 A0A023F553 A0A067R0I1 N6UC94 A0A195EZI6 A0A0N7Z8X6 D6WJR7 A0A158P2R1 A0A0P4VPQ7 A0A026VU91 A0A0C9Q259 A0A0A9X8N7 K7IXM6 E0VJR2 A0A0A9W7P1 A0A0J7KML9 E9IPJ7 A0A336K4Q7 F4W5S0 A0A195DGJ8 A0A3Q0J7Z6 A0A151X2K9 B4LDZ8 A0A0Q9WUW3 B4KYH7 B4J3N7 A0A1L8DGM0 A0A154PEV8 A0A0A1WYU9 A0A3B0K0A8 A0A034W076 W8BEZ4 A0A034W4C9 A0A0A1WLG9 B4H7T2 Q29D94 A0A1W4VMF8 B3NDQ2 B3M9T8 A0A0K8V083 A0A2A3ECH6 B4ITV4 U4UAW4 A0A1Z5LDW7 Q9VUX0 A0A0J9RW12 B4QLS5 A0A1Q3FCD0 V5IFU9 A0A0K8TR68 Q7YU53 M9PFN0 A0A023FJT2 A0A023GK94 G3MP86 J9JLI7 A0A2S2QHR5 A0A131XFW3 A0A131ZDT6 L7M156 A0A2S2PNP6 A0A2R5LJT7 A0A2H8TEC6 A0A293M2H0 A0A1I8JVR3 A0A2M4CPD0 A0A0K8RRI2 A0A1I8MRU9 B4N728 A0A210QY34 E9G5C4 A0A232F0E4 A0A023FYD6 A0A2R7X5F7 A0A1E1X2F2 C3YYP1 A0A224YV46 A0A1B0GCP0 B4HIM4 A0A162NJ95 A0A2C9JQ61
A0A1B6CPC4 A0A2P8Z7T5 A0A1W4XCY2 A0A1Y1KLL5 A0A0M8ZX50 A0A1B6JIN2 A0A1B6MJA2 A0A088AD85 E2BAZ1 A0A224XV12 A0A023F553 A0A067R0I1 N6UC94 A0A195EZI6 A0A0N7Z8X6 D6WJR7 A0A158P2R1 A0A0P4VPQ7 A0A026VU91 A0A0C9Q259 A0A0A9X8N7 K7IXM6 E0VJR2 A0A0A9W7P1 A0A0J7KML9 E9IPJ7 A0A336K4Q7 F4W5S0 A0A195DGJ8 A0A3Q0J7Z6 A0A151X2K9 B4LDZ8 A0A0Q9WUW3 B4KYH7 B4J3N7 A0A1L8DGM0 A0A154PEV8 A0A0A1WYU9 A0A3B0K0A8 A0A034W076 W8BEZ4 A0A034W4C9 A0A0A1WLG9 B4H7T2 Q29D94 A0A1W4VMF8 B3NDQ2 B3M9T8 A0A0K8V083 A0A2A3ECH6 B4ITV4 U4UAW4 A0A1Z5LDW7 Q9VUX0 A0A0J9RW12 B4QLS5 A0A1Q3FCD0 V5IFU9 A0A0K8TR68 Q7YU53 M9PFN0 A0A023FJT2 A0A023GK94 G3MP86 J9JLI7 A0A2S2QHR5 A0A131XFW3 A0A131ZDT6 L7M156 A0A2S2PNP6 A0A2R5LJT7 A0A2H8TEC6 A0A293M2H0 A0A1I8JVR3 A0A2M4CPD0 A0A0K8RRI2 A0A1I8MRU9 B4N728 A0A210QY34 E9G5C4 A0A232F0E4 A0A023FYD6 A0A2R7X5F7 A0A1E1X2F2 C3YYP1 A0A224YV46 A0A1B0GCP0 B4HIM4 A0A162NJ95 A0A2C9JQ61
PDB
3PGL
E-value=3.04961e-85,
Score=802
Ontologies
GO
PANTHER
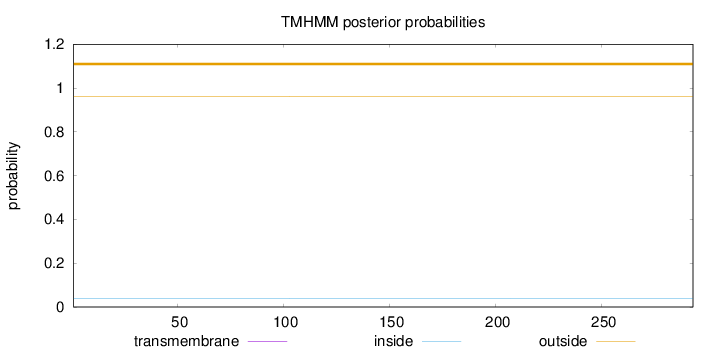
Topology
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000700000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03788
outside
1 - 293
Population Genetic Test Statistics
Pi
192.863314
Theta
161.623225
Tajima's D
0.02736
CLR
1118.833229
CSRT
0.381230938453077
Interpretation
Uncertain
