Pre Gene Modal
BGIBMGA006011
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101740551_[Bombyx_mori]
Full name
mRNA-capping enzyme
Alternative Name
HCE
MCE1
MCE1
Location in the cell
Nuclear Reliability : 4.343
Sequence
CDS
ATGATTTTCGACTTCTCTAAAAGTATTAAGAAAAAAATAGGTTTATGGATTGATCTCACAAATACAAGCAGATTTTACGATAAAGGGGAGATAGAGGAAATGGACTGTAAATATATGAAAATACAATGCAAAGGTCACGGAGAGGCACCGAGTGAAGACCAAATAAGGGTCTTCATACACACTGTCAAGAACTTTATGAAGAAAAAACCTCTTGAGCTCATAGCTGTTCACTGCACACATGGATTTAACAGAACCGGTTTCTTGATTGTGTCTTTCATGGTTCAAGAAATGGACTGTAGTATTGAAGCTGCCTTAAAAGAGTTTAATACAAAAAGACCTCCAGGAATATACAAGCAAGATTACCTCACTGAATTGTTCAAACTATATGGAGATGTTGAAGATACTCCAATGGCTCCTGCAAGACCTGACTGGTGCAATGAAATAGAAGAAGAAGAATCTGATGATGACAACCCGAGTGCTACACAGGATACATCAGAATGTAGCGACAGGAAACAGAAGAAAGGTAAACACAATAACATTATAAAGAAGGACTTTATGCCCGGCGTTAAGAATGTTGTTCCATTTGAGATTCAACCTCGAGTGAGTGAGATACAAAAGAAAGCACAGAAGTTCTGCCACTGGAAGAGCAAAGGCTTCCCCGGCTCGCAGCCCGTGTCGATGAACATGAACAACCTGTCGCTACTCAAGAAGCCGTACAGGGTCTCATGGAAAGCAGACGGCTTCAGATATATGATGTTAATAGACGGCGAAGACGAAGTGTACTTTTTGGACAGAGACAACTACGTATTCAAAGTTCACGGCCTCAAGTTTCTACATAAATCCGATCTGACGCATCACCTCAAGGACACTCTGCTCGATGGTGAGATGGTCATAGACAAGCAGAACGGCATTGAAACGCCGAGGTACTTGTGTTATGACATCATAGCCTTCGAGAACGAAGACATCGGAAAAGCTGAATTCTACCCGACAAGACTGAACTACATAAGTGAAGAAATCATCAATCCCAGGATACGGGCAATGACCAAAGGCTTAATAAGGAGAGAGAAGGAGCCATTCGGTGTGAGACGAAAAGAGTTTTGGGACATCACGATGTCGAGGCAACTACTCGGCGAGAAGTTCGCCAGGCAACTCCTCCACGAACCAGACGGGCTCATATTTCAGCCCTCTCTTGAGCCGTACATCGCTGGCACCTGCGACAAAGTGCTGAAATGGAAACCCGGTCACATGAACAGTTTAGATTTTCTGCTCAAAATCGACACTGAAAGCGGAGTGGGTATCGTTAGGAAAAAAATTGGACTATTGTACGTTGGACAGTGCTCGAACCCATGGGACACAATAAAGCTGACGAAAGAATTAAAAAATTATGACAACAAGATAATAGAATGTAAGTTCGAGAATAATCAGTGGGTGTTCATGCGAGAGCGGACGGACAAGTCCTTCCCCAACAGCCAGAACACGGCCATGGCCGTTTGGGAATCGATCAAGAACCCCGTCACTAAAGAGATTCTGCTGCATTACATCGACAAAAAGAGATTCATTAATGACGGATCCGAATCCGGTGCGCCGCCCAGCAAAATGGCTCGCACCATGTCCGATCAATAG
Protein
MIFDFSKSIKKKIGLWIDLTNTSRFYDKGEIEEMDCKYMKIQCKGHGEAPSEDQIRVFIHTVKNFMKKKPLELIAVHCTHGFNRTGFLIVSFMVQEMDCSIEAALKEFNTKRPPGIYKQDYLTELFKLYGDVEDTPMAPARPDWCNEIEEEESDDDNPSATQDTSECSDRKQKKGKHNNIIKKDFMPGVKNVVPFEIQPRVSEIQKKAQKFCHWKSKGFPGSQPVSMNMNNLSLLKKPYRVSWKADGFRYMMLIDGEDEVYFLDRDNYVFKVHGLKFLHKSDLTHHLKDTLLDGEMVIDKQNGIETPRYLCYDIIAFENEDIGKAEFYPTRLNYISEEIINPRIRAMTKGLIRREKEPFGVRRKEFWDITMSRQLLGEKFARQLLHEPDGLIFQPSLEPYIAGTCDKVLKWKPGHMNSLDFLLKIDTESGVGIVRKKIGLLYVGQCSNPWDTIKLTKELKNYDNKIIECKFENNQWVFMRERTDKSFPNSQNTAMAVWESIKNPVTKEILLHYIDKKRFINDGSESGAPPSKMARTMSDQ
Summary
Description
Bifunctional mRNA-capping enzyme exhibiting RNA 5'-triphosphatase activity in the N-terminal part and mRNA guanylyltransferase activity in the C-terminal part. Catalyzes the first two steps of cap formation: by removing the gamma-phosphate from the 5'-triphosphate end of nascent mRNA to yield a diphosphate end, and by transferring the gmp moiety of GTP to the 5'-diphosphate terminus.
Catalytic Activity
a 5'-diphospho-(purine-ribonucleotide)-[mRNA] + GTP + H(+) = a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + diphosphate
a 5'-triphospho-(purine-ribonucleotide)-[mRNA] + H2O = a 5'-diphospho-(purine-ribonucleotide)-[mRNA] + H(+) + phosphate
a 5'-triphospho-(purine-ribonucleotide)-[mRNA] + H2O = a 5'-diphospho-(purine-ribonucleotide)-[mRNA] + H(+) + phosphate
Subunit
Interacts with SUPT5H and RNMT (By similarity). Interacts with POLR2A (via C-terminus); this enhances guanylyltransferase activity. Binds (via GTase domain) to the elongating phosphorylated form of RNA polymerase II; can form direct interactions with the phosphorylated POLR2A C-terminal domain and indirect interactions via bound RNA.
Similarity
In the C-terminal section; belongs to the eukaryotic GTase family.
In the N-terminal section; belongs to the non-receptor class of the protein-tyrosine phosphatase family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
In the N-terminal section; belongs to the non-receptor class of the protein-tyrosine phosphatase family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Keywords
3D-structure
Complete proteome
GTP-binding
Hydrolase
mRNA capping
mRNA processing
Multifunctional enzyme
Nucleotide-binding
Nucleotidyltransferase
Nucleus
Reference proteome
Transferase
Feature
chain mRNA-capping enzyme
Uniprot
H9J919
A0A2H1VYM1
A0A2A4JQW3
A0A194Q0P7
A0A1Y1K3U6
A0A1W4X0Q2
+ More
A0A026WXY9 A0A0J7KVC7 D6WLP1 A0A336MHT3 A0A1Z5L9F7 A0A1L8DGW7 A0A067R5A4 A0A2R5LG19 E1ZZH4 A0A069DWC6 A0A293MZE1 J3JY60 A0A232ETW5 A0A0P4VTU9 F4WLL9 N6TAN6 A0A0M9A9B4 A0A0L7R5E6 K7IPT8 A0A0V0G8H0 A0A131XW43 A0A158NNC9 A0A088A1S3 A0A195EE52 A0A0Q9WN74 A0A0Q9WN75 T1HRH5 A0A154P0X3 A0A182N872 A0A182WAC7 T1ITR8 A0A1B6JT89 B0XGS4 A0A310SGT9 A0A131XBV8 A0A084W2G9 A0A182R1A0 J9KB51 A0A0C9R0J2 A0A182M5H2 W8BUE4 Q9VY44 A0A2C9K4I8 A0A131YIZ2 A0A0P4ZTT3 A0A182IUG8 A0A182P5A2 L7M2G5 A0A2S2QMC2 B4IGB9 A0A0P6FV68 A0A0P5TAF3 B4R4N6 A0A0P5UB79 A0A224Z3H3 L7LUN1 B4Q2R9 A0A1L8G346 Q9IA93 E2BXL6 Q6NWX1 F7D3D6 E9IVQ6 L7LWT8 S7MEP9 A0A2A3ELR4 F7CUI3 A0A091I3N6 E9GIK6 A0A0K8SC56 A0A034WF87 K7FW18 Q3UA94 O55236 K1QXN6 A0A1U7QTZ6 D3ZH30 A0A0P5G4N7 A0A2J7Q8M4 A0A0A9WYI1 A0A0P6IFP2 A0A3P8YN88 W5NJX5 G3T193 A0A182YNQ8 Q3UCK1 A0A3P8YLJ9 G3W8W3 R0K8M5 U3J5W2 A0A3P9A8D0 W5PTQ0 A0A091INL4 A0A091W2Q2
A0A026WXY9 A0A0J7KVC7 D6WLP1 A0A336MHT3 A0A1Z5L9F7 A0A1L8DGW7 A0A067R5A4 A0A2R5LG19 E1ZZH4 A0A069DWC6 A0A293MZE1 J3JY60 A0A232ETW5 A0A0P4VTU9 F4WLL9 N6TAN6 A0A0M9A9B4 A0A0L7R5E6 K7IPT8 A0A0V0G8H0 A0A131XW43 A0A158NNC9 A0A088A1S3 A0A195EE52 A0A0Q9WN74 A0A0Q9WN75 T1HRH5 A0A154P0X3 A0A182N872 A0A182WAC7 T1ITR8 A0A1B6JT89 B0XGS4 A0A310SGT9 A0A131XBV8 A0A084W2G9 A0A182R1A0 J9KB51 A0A0C9R0J2 A0A182M5H2 W8BUE4 Q9VY44 A0A2C9K4I8 A0A131YIZ2 A0A0P4ZTT3 A0A182IUG8 A0A182P5A2 L7M2G5 A0A2S2QMC2 B4IGB9 A0A0P6FV68 A0A0P5TAF3 B4R4N6 A0A0P5UB79 A0A224Z3H3 L7LUN1 B4Q2R9 A0A1L8G346 Q9IA93 E2BXL6 Q6NWX1 F7D3D6 E9IVQ6 L7LWT8 S7MEP9 A0A2A3ELR4 F7CUI3 A0A091I3N6 E9GIK6 A0A0K8SC56 A0A034WF87 K7FW18 Q3UA94 O55236 K1QXN6 A0A1U7QTZ6 D3ZH30 A0A0P5G4N7 A0A2J7Q8M4 A0A0A9WYI1 A0A0P6IFP2 A0A3P8YN88 W5NJX5 G3T193 A0A182YNQ8 Q3UCK1 A0A3P8YLJ9 G3W8W3 R0K8M5 U3J5W2 A0A3P9A8D0 W5PTQ0 A0A091INL4 A0A091W2Q2
Pubmed
19121390
26354079
28004739
24508170
30249741
18362917
+ More
19820115 28528879 24845553 20798317 26334808 22516182 28648823 27129103 21719571 23537049 20075255 29652888 21347285 17994087 28049606 24438588 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15562597 26830274 25576852 28797301 17550304 27762356 10679253 20431018 21282665 18464734 21292972 25348373 17381049 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 9371772 9407024 15489334 9770468 11350947 21683636 22992520 15057822 15632090 25401762 26823975 25069045 25244985 21709235 23749191 20809919
19820115 28528879 24845553 20798317 26334808 22516182 28648823 27129103 21719571 23537049 20075255 29652888 21347285 17994087 28049606 24438588 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15562597 26830274 25576852 28797301 17550304 27762356 10679253 20431018 21282665 18464734 21292972 25348373 17381049 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 9371772 9407024 15489334 9770468 11350947 21683636 22992520 15057822 15632090 25401762 26823975 25069045 25244985 21709235 23749191 20809919
EMBL
BABH01004644
BABH01004645
ODYU01005241
SOQ45907.1
NWSH01000867
PCG73800.1
+ More
KQ459584 KPI98564.1 GEZM01093755 JAV56144.1 KK107085 QOIP01000008 EZA60009.1 RLU19773.1 LBMM01002876 KMQ94258.1 KQ971343 EFA03427.1 UFQT01001020 SSX28519.1 GFJQ02002933 JAW04037.1 GFDF01008457 JAV05627.1 KK852692 KDR18340.1 GGLE01004337 MBY08463.1 GL435347 EFN73425.1 GBGD01000907 JAC87982.1 GFWV01021483 GFWV01021484 MAA46212.1 BT128184 AEE63145.1 NNAY01002212 OXU21792.1 GDKW01000525 JAI56070.1 GL888208 EGI64955.1 APGK01037499 KB740948 KB632288 ENN77339.1 ERL91515.1 KQ435710 KOX79850.1 KQ414652 KOC66102.1 GECL01001740 JAP04384.1 GEFM01005368 GEGO01001496 JAP70428.1 JAR93908.1 ADTU01021137 KQ979039 KYN23386.1 CH933810 KRF94242.1 KRF94241.1 ACPB03014947 KQ434792 KZC05483.1 JH431494 GECU01005268 JAT02439.1 DS233060 EDS27797.1 KQ764881 OAD54353.1 GEFH01004749 JAP63832.1 ATLV01019619 KE525275 KFB44413.1 AXCN02001004 ABLF02031184 GBYB01000346 JAG70113.1 AXCM01000427 GAMC01009624 JAB96931.1 AE014298 BT009965 AAF48361.1 AAQ22434.1 GEDV01009630 JAP78927.1 GDIP01211639 LRGB01000752 JAJ11763.1 KZS16162.1 GACK01006799 JAA58235.1 GGMS01009702 MBY78905.1 CH480835 EDW48853.1 GDIQ01043923 JAN50814.1 GDIP01130838 JAL72876.1 CM000366 EDX17890.1 GDIP01119187 JAL84527.1 GFPF01009967 MAA21113.1 GACK01009722 JAA55312.1 CM000162 EDX01663.2 CM004475 OCT78282.1 AF218793 BC169624 AAF43143.1 AAI69624.1 GL451265 EFN79585.1 BC067387 AAH67387.1 AAMC01108638 AAMC01108639 AAMC01108640 AAMC01108641 AAMC01108642 AAMC01108643 AAMC01108644 AAMC01108645 AAMC01108646 AAMC01108647 GL766328 EFZ15363.1 GACK01009725 JAA55309.1 KE161261 EPQ02804.1 KZ288215 PBC32733.1 KL218085 KFP02083.1 GL732546 EFX80737.1 GBRD01014939 JAG50887.1 GAKP01005583 JAC53369.1 AGCU01154571 AGCU01154572 AGCU01154573 AGCU01154574 AGCU01154575 AGCU01154576 AGCU01154577 AGCU01154578 AGCU01154579 AGCU01154580 AK142016 AK151463 AK160007 AK169305 CH466538 BAE24914.1 BAE30421.1 EDL05484.1 AF025653 AF034568 BC043657 JH819141 EKC33700.1 AABR07047801 AABR07047802 AABR07047803 AABR07047804 CH473962 EDL98586.1 GDIQ01246340 JAK05385.1 NEVH01016950 PNF24935.1 GBHO01030082 GBRD01014938 GBRD01014937 GDHC01017906 JAG13522.1 JAG50888.1 JAQ00723.1 GDIQ01005240 JAN89497.1 AHAT01011937 AHAT01011938 AHAT01011939 AHAT01011940 AHAT01011941 AHAT01011942 AK150497 BAE29611.1 AEFK01169123 AEFK01169124 AEFK01169125 AEFK01169126 AEFK01169127 AEFK01169128 AEFK01169129 AEFK01169130 AEFK01169131 AEFK01169132 KB742594 EOB06601.1 ADON01023818 ADON01023819 ADON01023820 ADON01023821 ADON01023822 ADON01023823 ADON01023824 ADON01023825 ADON01023826 ADON01023827 AMGL01107335 AMGL01107336 AMGL01107337 AMGL01107338 AMGL01107339 AMGL01107340 AMGL01107341 KK500574 KFP08980.1 KL411541 KFR09330.1
KQ459584 KPI98564.1 GEZM01093755 JAV56144.1 KK107085 QOIP01000008 EZA60009.1 RLU19773.1 LBMM01002876 KMQ94258.1 KQ971343 EFA03427.1 UFQT01001020 SSX28519.1 GFJQ02002933 JAW04037.1 GFDF01008457 JAV05627.1 KK852692 KDR18340.1 GGLE01004337 MBY08463.1 GL435347 EFN73425.1 GBGD01000907 JAC87982.1 GFWV01021483 GFWV01021484 MAA46212.1 BT128184 AEE63145.1 NNAY01002212 OXU21792.1 GDKW01000525 JAI56070.1 GL888208 EGI64955.1 APGK01037499 KB740948 KB632288 ENN77339.1 ERL91515.1 KQ435710 KOX79850.1 KQ414652 KOC66102.1 GECL01001740 JAP04384.1 GEFM01005368 GEGO01001496 JAP70428.1 JAR93908.1 ADTU01021137 KQ979039 KYN23386.1 CH933810 KRF94242.1 KRF94241.1 ACPB03014947 KQ434792 KZC05483.1 JH431494 GECU01005268 JAT02439.1 DS233060 EDS27797.1 KQ764881 OAD54353.1 GEFH01004749 JAP63832.1 ATLV01019619 KE525275 KFB44413.1 AXCN02001004 ABLF02031184 GBYB01000346 JAG70113.1 AXCM01000427 GAMC01009624 JAB96931.1 AE014298 BT009965 AAF48361.1 AAQ22434.1 GEDV01009630 JAP78927.1 GDIP01211639 LRGB01000752 JAJ11763.1 KZS16162.1 GACK01006799 JAA58235.1 GGMS01009702 MBY78905.1 CH480835 EDW48853.1 GDIQ01043923 JAN50814.1 GDIP01130838 JAL72876.1 CM000366 EDX17890.1 GDIP01119187 JAL84527.1 GFPF01009967 MAA21113.1 GACK01009722 JAA55312.1 CM000162 EDX01663.2 CM004475 OCT78282.1 AF218793 BC169624 AAF43143.1 AAI69624.1 GL451265 EFN79585.1 BC067387 AAH67387.1 AAMC01108638 AAMC01108639 AAMC01108640 AAMC01108641 AAMC01108642 AAMC01108643 AAMC01108644 AAMC01108645 AAMC01108646 AAMC01108647 GL766328 EFZ15363.1 GACK01009725 JAA55309.1 KE161261 EPQ02804.1 KZ288215 PBC32733.1 KL218085 KFP02083.1 GL732546 EFX80737.1 GBRD01014939 JAG50887.1 GAKP01005583 JAC53369.1 AGCU01154571 AGCU01154572 AGCU01154573 AGCU01154574 AGCU01154575 AGCU01154576 AGCU01154577 AGCU01154578 AGCU01154579 AGCU01154580 AK142016 AK151463 AK160007 AK169305 CH466538 BAE24914.1 BAE30421.1 EDL05484.1 AF025653 AF034568 BC043657 JH819141 EKC33700.1 AABR07047801 AABR07047802 AABR07047803 AABR07047804 CH473962 EDL98586.1 GDIQ01246340 JAK05385.1 NEVH01016950 PNF24935.1 GBHO01030082 GBRD01014938 GBRD01014937 GDHC01017906 JAG13522.1 JAG50888.1 JAQ00723.1 GDIQ01005240 JAN89497.1 AHAT01011937 AHAT01011938 AHAT01011939 AHAT01011940 AHAT01011941 AHAT01011942 AK150497 BAE29611.1 AEFK01169123 AEFK01169124 AEFK01169125 AEFK01169126 AEFK01169127 AEFK01169128 AEFK01169129 AEFK01169130 AEFK01169131 AEFK01169132 KB742594 EOB06601.1 ADON01023818 ADON01023819 ADON01023820 ADON01023821 ADON01023822 ADON01023823 ADON01023824 ADON01023825 ADON01023826 ADON01023827 AMGL01107335 AMGL01107336 AMGL01107337 AMGL01107338 AMGL01107339 AMGL01107340 AMGL01107341 KK500574 KFP08980.1 KL411541 KFR09330.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000192223
UP000053097
UP000279307
+ More
UP000036403 UP000007266 UP000027135 UP000000311 UP000215335 UP000007755 UP000019118 UP000030742 UP000053105 UP000053825 UP000002358 UP000005205 UP000005203 UP000078492 UP000009192 UP000015103 UP000076502 UP000075884 UP000075920 UP000002320 UP000030765 UP000075886 UP000007819 UP000075883 UP000000803 UP000076420 UP000076858 UP000075880 UP000075885 UP000001292 UP000000304 UP000002282 UP000186698 UP000008237 UP000008143 UP000242457 UP000002279 UP000054308 UP000000305 UP000007267 UP000000589 UP000005408 UP000189706 UP000002494 UP000235965 UP000265140 UP000018468 UP000007646 UP000076408 UP000007648 UP000016666 UP000002356 UP000053119 UP000053283
UP000036403 UP000007266 UP000027135 UP000000311 UP000215335 UP000007755 UP000019118 UP000030742 UP000053105 UP000053825 UP000002358 UP000005205 UP000005203 UP000078492 UP000009192 UP000015103 UP000076502 UP000075884 UP000075920 UP000002320 UP000030765 UP000075886 UP000007819 UP000075883 UP000000803 UP000076420 UP000076858 UP000075880 UP000075885 UP000001292 UP000000304 UP000002282 UP000186698 UP000008237 UP000008143 UP000242457 UP000002279 UP000054308 UP000000305 UP000007267 UP000000589 UP000005408 UP000189706 UP000002494 UP000235965 UP000265140 UP000018468 UP000007646 UP000076408 UP000007648 UP000016666 UP000002356 UP000053119 UP000053283
Pfam
Interpro
IPR011538
Nuo51_FMN-bd
+ More
IPR029021 Prot-tyrosine_phosphatase-like
IPR019554 Soluble_ligand-bd
IPR011537 NADH-UbQ_OxRdtase_suF
IPR037225 Nuo51_FMN-bd_sf
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR001949 NADH-UbQ_OxRdtase_51kDa_CS
IPR001339 mRNA_cap_enzyme
IPR000340 Dual-sp_phosphatase_cat-dom
IPR017074 mRNA_cap_enz_bifunc
IPR012340 NA-bd_OB-fold
IPR013846 mRNA_cap_enzyme_C
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR027417 P-loop_NTPase
IPR036961 Kinesin_motor_dom_sf
IPR001609 Myosin_head_motor_dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR019554 Soluble_ligand-bd
IPR011537 NADH-UbQ_OxRdtase_suF
IPR037225 Nuo51_FMN-bd_sf
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR001949 NADH-UbQ_OxRdtase_51kDa_CS
IPR001339 mRNA_cap_enzyme
IPR000340 Dual-sp_phosphatase_cat-dom
IPR017074 mRNA_cap_enz_bifunc
IPR012340 NA-bd_OB-fold
IPR013846 mRNA_cap_enzyme_C
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR027417 P-loop_NTPase
IPR036961 Kinesin_motor_dom_sf
IPR001609 Myosin_head_motor_dom
SUPFAM
ProteinModelPortal
H9J919
A0A2H1VYM1
A0A2A4JQW3
A0A194Q0P7
A0A1Y1K3U6
A0A1W4X0Q2
+ More
A0A026WXY9 A0A0J7KVC7 D6WLP1 A0A336MHT3 A0A1Z5L9F7 A0A1L8DGW7 A0A067R5A4 A0A2R5LG19 E1ZZH4 A0A069DWC6 A0A293MZE1 J3JY60 A0A232ETW5 A0A0P4VTU9 F4WLL9 N6TAN6 A0A0M9A9B4 A0A0L7R5E6 K7IPT8 A0A0V0G8H0 A0A131XW43 A0A158NNC9 A0A088A1S3 A0A195EE52 A0A0Q9WN74 A0A0Q9WN75 T1HRH5 A0A154P0X3 A0A182N872 A0A182WAC7 T1ITR8 A0A1B6JT89 B0XGS4 A0A310SGT9 A0A131XBV8 A0A084W2G9 A0A182R1A0 J9KB51 A0A0C9R0J2 A0A182M5H2 W8BUE4 Q9VY44 A0A2C9K4I8 A0A131YIZ2 A0A0P4ZTT3 A0A182IUG8 A0A182P5A2 L7M2G5 A0A2S2QMC2 B4IGB9 A0A0P6FV68 A0A0P5TAF3 B4R4N6 A0A0P5UB79 A0A224Z3H3 L7LUN1 B4Q2R9 A0A1L8G346 Q9IA93 E2BXL6 Q6NWX1 F7D3D6 E9IVQ6 L7LWT8 S7MEP9 A0A2A3ELR4 F7CUI3 A0A091I3N6 E9GIK6 A0A0K8SC56 A0A034WF87 K7FW18 Q3UA94 O55236 K1QXN6 A0A1U7QTZ6 D3ZH30 A0A0P5G4N7 A0A2J7Q8M4 A0A0A9WYI1 A0A0P6IFP2 A0A3P8YN88 W5NJX5 G3T193 A0A182YNQ8 Q3UCK1 A0A3P8YLJ9 G3W8W3 R0K8M5 U3J5W2 A0A3P9A8D0 W5PTQ0 A0A091INL4 A0A091W2Q2
A0A026WXY9 A0A0J7KVC7 D6WLP1 A0A336MHT3 A0A1Z5L9F7 A0A1L8DGW7 A0A067R5A4 A0A2R5LG19 E1ZZH4 A0A069DWC6 A0A293MZE1 J3JY60 A0A232ETW5 A0A0P4VTU9 F4WLL9 N6TAN6 A0A0M9A9B4 A0A0L7R5E6 K7IPT8 A0A0V0G8H0 A0A131XW43 A0A158NNC9 A0A088A1S3 A0A195EE52 A0A0Q9WN74 A0A0Q9WN75 T1HRH5 A0A154P0X3 A0A182N872 A0A182WAC7 T1ITR8 A0A1B6JT89 B0XGS4 A0A310SGT9 A0A131XBV8 A0A084W2G9 A0A182R1A0 J9KB51 A0A0C9R0J2 A0A182M5H2 W8BUE4 Q9VY44 A0A2C9K4I8 A0A131YIZ2 A0A0P4ZTT3 A0A182IUG8 A0A182P5A2 L7M2G5 A0A2S2QMC2 B4IGB9 A0A0P6FV68 A0A0P5TAF3 B4R4N6 A0A0P5UB79 A0A224Z3H3 L7LUN1 B4Q2R9 A0A1L8G346 Q9IA93 E2BXL6 Q6NWX1 F7D3D6 E9IVQ6 L7LWT8 S7MEP9 A0A2A3ELR4 F7CUI3 A0A091I3N6 E9GIK6 A0A0K8SC56 A0A034WF87 K7FW18 Q3UA94 O55236 K1QXN6 A0A1U7QTZ6 D3ZH30 A0A0P5G4N7 A0A2J7Q8M4 A0A0A9WYI1 A0A0P6IFP2 A0A3P8YN88 W5NJX5 G3T193 A0A182YNQ8 Q3UCK1 A0A3P8YLJ9 G3W8W3 R0K8M5 U3J5W2 A0A3P9A8D0 W5PTQ0 A0A091INL4 A0A091W2Q2
PDB
3S24
E-value=7.08239e-100,
Score=931
Ontologies
GO
PANTHER
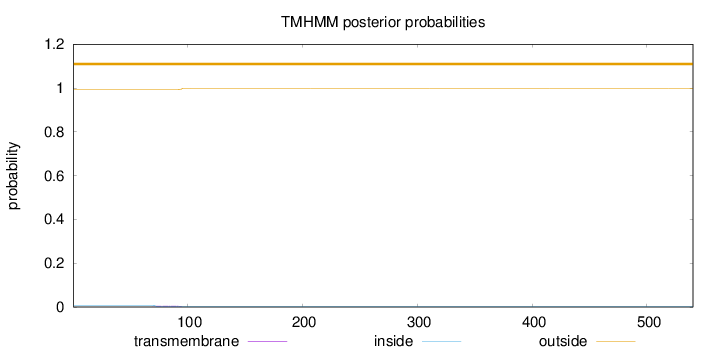
Topology
Subcellular location
Nucleus
Length:
540
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12329
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00690
outside
1 - 540
Population Genetic Test Statistics
Pi
21.968726
Theta
154.0176
Tajima's D
0.740743
CLR
1.652143
CSRT
0.584220788960552
Interpretation
Uncertain
