Pre Gene Modal
BGIBMGA006139
Annotation
PREDICTED:_beta-lactamase-like_protein_2_homolog_[Papilio_machaon]
Full name
Beta-lactamase-like protein 2 homolog
Location in the cell
Nuclear Reliability : 4.126
Sequence
CDS
ATGGCTGCTGTTATTCCACCAGTAACTAAATTATCCACATCTATTATTAGGATTCTCGGTTGTAATCCAGGTCCAATGACTCTTCAAGGCACAAACACGTATCTCATTGGTTCCGGTAAAAATCGAATCTTAGTTGACACTGGAGACAACAATATACCCGAGTACCAGAAACATTTATCAGATGTTGTGAACTCTGAAAAAGCTAACATAGAACATATTGTTCTCACTCACTGGCACCATGACCATGTTGGTGGTGTCGAGAACATATACAAAACTATAGCAAAAAATCCCATAATATGGAAGCACAAGAAGAGTTCAGAAGATTCTGCTGATAATGAACTACCAGCATCTATTCCAATAGAATGGCTTAGAGACGGACAAGAAATTAAAGTAGAAGGAGCTACAGTCAAAGTGCATCATACTCCAGGACACACAACTGACCACATAGTGTTGACATGTCTGGAAGAAAACATTTTATTCAGTGGCGATTGCATATTAGGAGAGGGGACAGCTGTGTTTGAAGACTTATACACCTATATGAAAAGTCTGCAAAAAATATTGGACTTAAAACCAAACATTATTTACCCTGGTCATGGGAATATAGTAAAGGAACCCGTTGAAAAAATTGAATATTATATAGCTCACAGAAATCAAAGAGAAGAACAAATATTAGAAGCTTTGAAAACAAATTCTGATAAAAATCTAAATGAAATGGATTTAGTGAAAATAATATACACCGAGACACCAGAACACTTGTGGCCAGCAGCTGCATATAATGTAAATCACCACTTGACCAAATTAGTTAAAGAATGCAAAGTTAAGTGTGTTAATAACGATGGTGAAAATAGATGGCAGTACATTTCAAAGACGTCTAGTAATTTATAA
Protein
MAAVIPPVTKLSTSIIRILGCNPGPMTLQGTNTYLIGSGKNRILVDTGDNNIPEYQKHLSDVVNSEKANIEHIVLTHWHHDHVGGVENIYKTIAKNPIIWKHKKSSEDSADNELPASIPIEWLRDGQEIKVEGATVKVHHTPGHTTDHIVLTCLEENILFSGDCILGEGTAVFEDLYTYMKSLQKILDLKPNIIYPGHGNIVKEPVEKIEYYIAHRNQREEQILEALKTNSDKNLNEMDLVKIIYTETPEHLWPAAAYNVNHHLTKLVKECKVKCVNNDGENRWQYISKTSSNL
Summary
Similarity
Belongs to the metallo-beta-lactamase superfamily. Glyoxalase II family.
Keywords
Complete proteome
Hydrolase
Metal-binding
Reference proteome
Zinc
Feature
chain Beta-lactamase-like protein 2 homolog
Uniprot
H9J9E7
A0A212EVV0
S4PSF0
A0A194R7K9
A0A194PZV4
A0A2A4JPQ6
+ More
A0A2H1VYX8 A0A1I8N9I2 A0A1I8N9G5 A0A0K8WLR5 A0A1I8PBI5 A0A2P8YPV6 A0A034WRD8 A0A1I8PBQ2 B4MA58 D3TQU0 B4L6Y6 A0A0Q9WD46 A0A0Q9XEZ4 A0A1A9ZVC9 A0A1A9UPI0 A0A1W4VWH8 A0A0L0CLD8 A0A1A9X4N4 B4MT92 A0A1A9XAI7 B4Q699 Q9VLS9 A0A0M3QZG8 B3N704 A0A1B0ATC1 B4HYE8 B3MMW9 B4JX40 B4NWD7 D6WLP4 V5I985 A0A182Y5D9 A0A182U9T5 A0A1Y1LCD1 W8C3H0 A0A1B0G451 A0A182MK55 A0A182Q8J6 A0A182XG46 A0A182L737 Q7QB16 A0A182VJ46 A0A182ID92 A0A3B0KR91 A0A1W4XBE1 A0A182SJX1 A0A084VBG8 B4HCY0 A0A2M3ZCD4 A0A182JPJ9 A0A2M4BVW8 W5JNB8 Q29CQ2 A0A182IQM7 A0A2M4BW28 A0A0T6ATM5 A0A023EPT1 A0A182G870 A0A2J7Q8M5 Q16K50 A0A1L8E520 A0A1L8E4T4 U5EIX2 A0A182FPT4 N6U6R6 A0A151WN88 A0A0C9Q1M0 A0A195DSL8 H2YXR9 A0A158NLS1 A0A195BKB2 A0A026WFQ2 A0A3L8DLJ2 A0A195FX76 A0A067R7A9 F4X2N9 A0A146NUU5 I3KCD2 A0A3B4G7H1 A0A3B3CGA4 A0A3P8N7X5 A0A3P9B6Z0 A0A154PM87 H2LML6 A0A3P9LIK6 A0A3B5PPQ3 A0A3B4XB33 A0A1W3JME3 A0A195CCQ0 A0A1S3I8Z0 E9IHH0 A0A182RV61 A0A3B4G627
A0A2H1VYX8 A0A1I8N9I2 A0A1I8N9G5 A0A0K8WLR5 A0A1I8PBI5 A0A2P8YPV6 A0A034WRD8 A0A1I8PBQ2 B4MA58 D3TQU0 B4L6Y6 A0A0Q9WD46 A0A0Q9XEZ4 A0A1A9ZVC9 A0A1A9UPI0 A0A1W4VWH8 A0A0L0CLD8 A0A1A9X4N4 B4MT92 A0A1A9XAI7 B4Q699 Q9VLS9 A0A0M3QZG8 B3N704 A0A1B0ATC1 B4HYE8 B3MMW9 B4JX40 B4NWD7 D6WLP4 V5I985 A0A182Y5D9 A0A182U9T5 A0A1Y1LCD1 W8C3H0 A0A1B0G451 A0A182MK55 A0A182Q8J6 A0A182XG46 A0A182L737 Q7QB16 A0A182VJ46 A0A182ID92 A0A3B0KR91 A0A1W4XBE1 A0A182SJX1 A0A084VBG8 B4HCY0 A0A2M3ZCD4 A0A182JPJ9 A0A2M4BVW8 W5JNB8 Q29CQ2 A0A182IQM7 A0A2M4BW28 A0A0T6ATM5 A0A023EPT1 A0A182G870 A0A2J7Q8M5 Q16K50 A0A1L8E520 A0A1L8E4T4 U5EIX2 A0A182FPT4 N6U6R6 A0A151WN88 A0A0C9Q1M0 A0A195DSL8 H2YXR9 A0A158NLS1 A0A195BKB2 A0A026WFQ2 A0A3L8DLJ2 A0A195FX76 A0A067R7A9 F4X2N9 A0A146NUU5 I3KCD2 A0A3B4G7H1 A0A3B3CGA4 A0A3P8N7X5 A0A3P9B6Z0 A0A154PM87 H2LML6 A0A3P9LIK6 A0A3B5PPQ3 A0A3B4XB33 A0A1W3JME3 A0A195CCQ0 A0A1S3I8Z0 E9IHH0 A0A182RV61 A0A3B4G627
EC Number
3.-.-.-
Pubmed
19121390
22118469
23622113
26354079
25315136
29403074
+ More
25348373 17994087 20353571 26108605 22936249 10731132 12537572 12537569 18057021 17550304 18362917 19820115 25244985 28004739 24495485 20966253 12364791 14747013 17210077 24438588 20920257 23761445 15632085 24945155 26483478 17510324 23537049 21347285 24508170 30249741 24845553 21719571 25186727 29451363 17554307 23542700 21282665
25348373 17994087 20353571 26108605 22936249 10731132 12537572 12537569 18057021 17550304 18362917 19820115 25244985 28004739 24495485 20966253 12364791 14747013 17210077 24438588 20920257 23761445 15632085 24945155 26483478 17510324 23537049 21347285 24508170 30249741 24845553 21719571 25186727 29451363 17554307 23542700 21282665
EMBL
BABH01004641
AGBW02012119
OWR45609.1
GAIX01013903
JAA78657.1
KQ460855
+ More
KPJ11856.1 KQ459584 KPI98563.1 NWSH01000867 PCG73799.1 ODYU01005241 SOQ45906.1 GDHF01000201 JAI52113.1 PYGN01000442 PSN46297.1 GAKP01002629 JAC56323.1 CH940655 EDW66117.1 EZ423792 ADD20068.1 CH933812 EDW06132.1 KRF82569.1 KRG07158.1 JRES01000325 KNC32264.1 CH963851 EDW75331.2 CM000361 CM002910 EDX04175.1 KMY88931.1 AE014134 AY122121 CP012528 ALC49349.1 CH954177 EDV59300.1 JXJN01003232 CH480818 EDW52078.1 CH902620 EDV30994.1 KPU73177.1 CH916376 EDV95316.1 CM000157 EDW88454.1 KRJ97806.1 KQ971343 EFA03426.1 GALX01003375 JAB65091.1 GEZM01064603 JAV68697.1 GAMC01006084 JAC00472.1 CCAG010008295 AXCM01001325 AXCN02001566 AAAB01008880 EAA08647.3 APCN01005054 OUUW01000011 SPP86428.1 ATLV01006809 KE524386 KFB35312.1 CH479512 EDW33359.1 GGFM01005453 MBW26204.1 GGFJ01008001 MBW57142.1 ADMH02000868 ETN64808.1 CH475434 EAL29289.2 GGFJ01008000 MBW57141.1 LJIG01022821 KRT78544.1 GAPW01003074 JAC10524.1 JXUM01047496 KQ561521 KXJ78299.1 NEVH01016950 PNF24938.1 CH477976 EAT34682.1 GFDF01000439 JAV13645.1 GFDF01000440 JAV13644.1 GANO01002462 JAB57409.1 APGK01037499 APGK01037500 KB740948 KB632288 ENN77340.1 ERL91513.1 KQ982919 KYQ49267.1 GBYB01007793 JAG77560.1 KQ980489 KYN15843.1 ADTU01019759 KQ976450 KYM85710.1 KK107238 EZA54763.1 QOIP01000007 RLU20779.1 KQ981219 KYN44449.1 KK852692 KDR18341.1 GL888591 EGI59235.1 GCES01151085 JAQ35237.1 AERX01017649 KQ434978 KZC12995.1 KQ977957 KYM98490.1 GL763279 EFZ19979.1
KPJ11856.1 KQ459584 KPI98563.1 NWSH01000867 PCG73799.1 ODYU01005241 SOQ45906.1 GDHF01000201 JAI52113.1 PYGN01000442 PSN46297.1 GAKP01002629 JAC56323.1 CH940655 EDW66117.1 EZ423792 ADD20068.1 CH933812 EDW06132.1 KRF82569.1 KRG07158.1 JRES01000325 KNC32264.1 CH963851 EDW75331.2 CM000361 CM002910 EDX04175.1 KMY88931.1 AE014134 AY122121 CP012528 ALC49349.1 CH954177 EDV59300.1 JXJN01003232 CH480818 EDW52078.1 CH902620 EDV30994.1 KPU73177.1 CH916376 EDV95316.1 CM000157 EDW88454.1 KRJ97806.1 KQ971343 EFA03426.1 GALX01003375 JAB65091.1 GEZM01064603 JAV68697.1 GAMC01006084 JAC00472.1 CCAG010008295 AXCM01001325 AXCN02001566 AAAB01008880 EAA08647.3 APCN01005054 OUUW01000011 SPP86428.1 ATLV01006809 KE524386 KFB35312.1 CH479512 EDW33359.1 GGFM01005453 MBW26204.1 GGFJ01008001 MBW57142.1 ADMH02000868 ETN64808.1 CH475434 EAL29289.2 GGFJ01008000 MBW57141.1 LJIG01022821 KRT78544.1 GAPW01003074 JAC10524.1 JXUM01047496 KQ561521 KXJ78299.1 NEVH01016950 PNF24938.1 CH477976 EAT34682.1 GFDF01000439 JAV13645.1 GFDF01000440 JAV13644.1 GANO01002462 JAB57409.1 APGK01037499 APGK01037500 KB740948 KB632288 ENN77340.1 ERL91513.1 KQ982919 KYQ49267.1 GBYB01007793 JAG77560.1 KQ980489 KYN15843.1 ADTU01019759 KQ976450 KYM85710.1 KK107238 EZA54763.1 QOIP01000007 RLU20779.1 KQ981219 KYN44449.1 KK852692 KDR18341.1 GL888591 EGI59235.1 GCES01151085 JAQ35237.1 AERX01017649 KQ434978 KZC12995.1 KQ977957 KYM98490.1 GL763279 EFZ19979.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000095301
+ More
UP000095300 UP000245037 UP000008792 UP000009192 UP000092445 UP000078200 UP000192221 UP000037069 UP000091820 UP000007798 UP000092443 UP000000304 UP000000803 UP000092553 UP000008711 UP000092460 UP000001292 UP000007801 UP000001070 UP000002282 UP000007266 UP000076408 UP000075902 UP000092444 UP000075883 UP000075886 UP000076407 UP000075882 UP000007062 UP000075903 UP000075840 UP000268350 UP000192223 UP000075901 UP000030765 UP000008744 UP000075881 UP000000673 UP000001819 UP000075880 UP000069940 UP000249989 UP000235965 UP000008820 UP000069272 UP000019118 UP000030742 UP000075809 UP000078492 UP000007875 UP000005205 UP000078540 UP000053097 UP000279307 UP000078541 UP000027135 UP000007755 UP000005207 UP000261460 UP000261560 UP000265100 UP000265160 UP000076502 UP000001038 UP000265180 UP000002852 UP000261360 UP000078542 UP000085678 UP000075900
UP000095300 UP000245037 UP000008792 UP000009192 UP000092445 UP000078200 UP000192221 UP000037069 UP000091820 UP000007798 UP000092443 UP000000304 UP000000803 UP000092553 UP000008711 UP000092460 UP000001292 UP000007801 UP000001070 UP000002282 UP000007266 UP000076408 UP000075902 UP000092444 UP000075883 UP000075886 UP000076407 UP000075882 UP000007062 UP000075903 UP000075840 UP000268350 UP000192223 UP000075901 UP000030765 UP000008744 UP000075881 UP000000673 UP000001819 UP000075880 UP000069940 UP000249989 UP000235965 UP000008820 UP000069272 UP000019118 UP000030742 UP000075809 UP000078492 UP000007875 UP000005205 UP000078540 UP000053097 UP000279307 UP000078541 UP000027135 UP000007755 UP000005207 UP000261460 UP000261560 UP000265100 UP000265160 UP000076502 UP000001038 UP000265180 UP000002852 UP000261360 UP000078542 UP000085678 UP000075900
Interpro
IPR036388
WH-like_DNA-bd_sf
+ More
IPR001279 Metallo-B-lactamas
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR041516 LACTB2_WH
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR028318 DYRK1A/B_MNB
IPR020683 Ankyrin_rpt-contain_dom
IPR013026 TPR-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR009060 UBA-like_sf
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR027417 P-loop_NTPase
IPR029071 Ubiquitin-like_domsf
IPR002110 Ankyrin_rpt
IPR015940 UBA
IPR001279 Metallo-B-lactamas
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR041516 LACTB2_WH
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR028318 DYRK1A/B_MNB
IPR020683 Ankyrin_rpt-contain_dom
IPR013026 TPR-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR009060 UBA-like_sf
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR027417 P-loop_NTPase
IPR029071 Ubiquitin-like_domsf
IPR002110 Ankyrin_rpt
IPR015940 UBA
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J9E7
A0A212EVV0
S4PSF0
A0A194R7K9
A0A194PZV4
A0A2A4JPQ6
+ More
A0A2H1VYX8 A0A1I8N9I2 A0A1I8N9G5 A0A0K8WLR5 A0A1I8PBI5 A0A2P8YPV6 A0A034WRD8 A0A1I8PBQ2 B4MA58 D3TQU0 B4L6Y6 A0A0Q9WD46 A0A0Q9XEZ4 A0A1A9ZVC9 A0A1A9UPI0 A0A1W4VWH8 A0A0L0CLD8 A0A1A9X4N4 B4MT92 A0A1A9XAI7 B4Q699 Q9VLS9 A0A0M3QZG8 B3N704 A0A1B0ATC1 B4HYE8 B3MMW9 B4JX40 B4NWD7 D6WLP4 V5I985 A0A182Y5D9 A0A182U9T5 A0A1Y1LCD1 W8C3H0 A0A1B0G451 A0A182MK55 A0A182Q8J6 A0A182XG46 A0A182L737 Q7QB16 A0A182VJ46 A0A182ID92 A0A3B0KR91 A0A1W4XBE1 A0A182SJX1 A0A084VBG8 B4HCY0 A0A2M3ZCD4 A0A182JPJ9 A0A2M4BVW8 W5JNB8 Q29CQ2 A0A182IQM7 A0A2M4BW28 A0A0T6ATM5 A0A023EPT1 A0A182G870 A0A2J7Q8M5 Q16K50 A0A1L8E520 A0A1L8E4T4 U5EIX2 A0A182FPT4 N6U6R6 A0A151WN88 A0A0C9Q1M0 A0A195DSL8 H2YXR9 A0A158NLS1 A0A195BKB2 A0A026WFQ2 A0A3L8DLJ2 A0A195FX76 A0A067R7A9 F4X2N9 A0A146NUU5 I3KCD2 A0A3B4G7H1 A0A3B3CGA4 A0A3P8N7X5 A0A3P9B6Z0 A0A154PM87 H2LML6 A0A3P9LIK6 A0A3B5PPQ3 A0A3B4XB33 A0A1W3JME3 A0A195CCQ0 A0A1S3I8Z0 E9IHH0 A0A182RV61 A0A3B4G627
A0A2H1VYX8 A0A1I8N9I2 A0A1I8N9G5 A0A0K8WLR5 A0A1I8PBI5 A0A2P8YPV6 A0A034WRD8 A0A1I8PBQ2 B4MA58 D3TQU0 B4L6Y6 A0A0Q9WD46 A0A0Q9XEZ4 A0A1A9ZVC9 A0A1A9UPI0 A0A1W4VWH8 A0A0L0CLD8 A0A1A9X4N4 B4MT92 A0A1A9XAI7 B4Q699 Q9VLS9 A0A0M3QZG8 B3N704 A0A1B0ATC1 B4HYE8 B3MMW9 B4JX40 B4NWD7 D6WLP4 V5I985 A0A182Y5D9 A0A182U9T5 A0A1Y1LCD1 W8C3H0 A0A1B0G451 A0A182MK55 A0A182Q8J6 A0A182XG46 A0A182L737 Q7QB16 A0A182VJ46 A0A182ID92 A0A3B0KR91 A0A1W4XBE1 A0A182SJX1 A0A084VBG8 B4HCY0 A0A2M3ZCD4 A0A182JPJ9 A0A2M4BVW8 W5JNB8 Q29CQ2 A0A182IQM7 A0A2M4BW28 A0A0T6ATM5 A0A023EPT1 A0A182G870 A0A2J7Q8M5 Q16K50 A0A1L8E520 A0A1L8E4T4 U5EIX2 A0A182FPT4 N6U6R6 A0A151WN88 A0A0C9Q1M0 A0A195DSL8 H2YXR9 A0A158NLS1 A0A195BKB2 A0A026WFQ2 A0A3L8DLJ2 A0A195FX76 A0A067R7A9 F4X2N9 A0A146NUU5 I3KCD2 A0A3B4G7H1 A0A3B3CGA4 A0A3P8N7X5 A0A3P9B6Z0 A0A154PM87 H2LML6 A0A3P9LIK6 A0A3B5PPQ3 A0A3B4XB33 A0A1W3JME3 A0A195CCQ0 A0A1S3I8Z0 E9IHH0 A0A182RV61 A0A3B4G627
PDB
4AD9
E-value=6.9196e-69,
Score=660
Ontologies
PANTHER
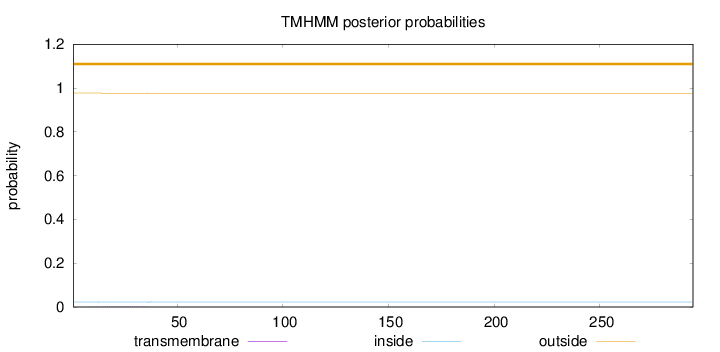
Topology
Length:
294
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03699
Exp number, first 60 AAs:
0.0364
Total prob of N-in:
0.02307
outside
1 - 294
Population Genetic Test Statistics
Pi
255.10574
Theta
209.43153
Tajima's D
0.672293
CLR
0.236932
CSRT
0.563321833908305
Interpretation
Uncertain
