Gene
KWMTBOMO01752 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006013
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101740551_[Bombyx_mori]
Full name
NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
Alternative Name
Complex I-51kD
NADH-ubiquinone oxidoreductase 51 kDa subunit
NADH-ubiquinone oxidoreductase 51 kDa subunit
Location in the cell
Nuclear Reliability : 4.319
Sequence
CDS
ATGGCTGGTGCACTGACAAGAGTGATTCAGGGCACAAAACCACATTTGGGCATTATTGGGCCTTTGGCAATTAATGTCAACAATGTACCGGTGCGCTTCCAGCAGACCCAGGCGCCTTCAAAAGACAAGTATGGACCGCTGGCCGATAGCGATCGGGTTTTCACAAACTTGTATGGTCGACATGAATGGAGGTTGAAGGGTGCTCTCGCCCGAGGGGACTGGTATTTGACGAAGGAGATTTTACTGAAAGGAACCGACTGGATCGTTAATGAAATGAAAACATCTGGCCTTCGAGGCAGGGGAGGAGCAGGTTTTCCAACAGGAATGAAGTGGTCATTCATGAACAAGCCATCAGATGGCCGTCCAAAATATCTCGTGGTGAATGCTGATGAGGGTGAACCTGGTACTTGTAAAGACAGGGAGATTATGCGTCACGACCCCCACAAACTGGTGGAAGGATGTCTAATTGCTGGCCGAGCCATGGGTGCACAAGCAGCATACATTTACATCAGAGGTGAATTCTACAATGAAGCAAGCAATTTGCAAGTTGCTATTGCTGAGGCTTACCAAGCCGGTCTCATTGGTAAGAACTCTTGTGGTTCTGGTTATGACTTTGATATATTCGTACACCGCGGTGCCGGCGCATACATCTGTGGTGAGGAGACAGCTCTCATTGAGTCCATTGAGGGCAAACAAGGCAAGCCGAGGCTCAAACCTCCTTTCCCCGCTGACGTGGGTTTGTTCGGTTGTCCCACCACTGTCAACAATGTAGAAACAATCGCGGTAGCGCCCACGATCTGTAGGCGTGGCGGCAAGTGGTTCGCATCGTTCGGGCGCCAACGTAACTCTGGCACGAAGCTGTTCAATATATCGGGTCACGTGAACACGCCGTGCACCGTGGAGGAGGAGATGAGCATCCCGCTCAAGGAGCTCATTGAGAGGCACGCGGGCGGAGTCACCGGCGGCTGGGACAACCTGCTCGCCATCATACCAGGAGGCTCCTCGACCCCGCTCATACCCAAGAACGTCTGCGAGACAGTGTTGATGGACTTCGACGGTCTAGTAGCCGCGCAGACCTCCCTGGGCACGGCGGCCATCATTGTCATGGACAAGTCAACTGACATCGTGAAGGCGATCGCTCGACTCATCATGTTCTACAAGCACGAGTCCTGCGGCCAGTGCACTCCTTGCCGCGAAGGTGTCTCCTGGATGAACAAGATTATATACAGATTCGTCGATGGTAATGCTTCACCGAAAGAAATCGACATGCTGTGGGAAATTTCGAAACAAATCGAAGGGCACACGATCTGCGCTCTGGGCGACGGAGCCGCGTGGCCGGTGCAGGGACTCATACGTCACTTCCGGCCCGAGCTCGAGCGACGCATGCAGGAGTACTCGGCCGCGCACGGGCCGAGCAAAGCCGAGCGCCTGTACTAG
Protein
MAGALTRVIQGTKPHLGIIGPLAINVNNVPVRFQQTQAPSKDKYGPLADSDRVFTNLYGRHEWRLKGALARGDWYLTKEILLKGTDWIVNEMKTSGLRGRGGAGFPTGMKWSFMNKPSDGRPKYLVVNADEGEPGTCKDREIMRHDPHKLVEGCLIAGRAMGAQAAYIYIRGEFYNEASNLQVAIAEAYQAGLIGKNSCGSGYDFDIFVHRGAGAYICGEETALIESIEGKQGKPRLKPPFPADVGLFGCPTTVNNVETIAVAPTICRRGGKWFASFGRQRNSGTKLFNISGHVNTPCTVEEEMSIPLKELIERHAGGVTGGWDNLLAIIPGGSSTPLIPKNVCETVLMDFDGLVAAQTSLGTAAIIVMDKSTDIVKAIARLIMFYKHESCGQCTPCREGVSWMNKIIYRFVDGNASPKEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELERRMQEYSAAHGPSKAERLY
Summary
Description
Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain.
Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
Catalytic Activity
A + H(+) + NADH = AH2 + NAD(+)
a ubiquinone + 5 H(+)(in) + NADH = a ubiquinol + 4 H(+)(out) + NAD(+)
a ubiquinone + 5 H(+)(in) + NADH = a ubiquinol + 4 H(+)(out) + NAD(+)
Cofactor
FMN
[4Fe-4S] cluster
[4Fe-4S] cluster
Subunit
Complex I is composed of 45 different subunits. This is a component of the flavoprotein-sulfur (FP) fragment of the enzyme (By similarity).
Similarity
Belongs to the complex I 51 kDa subunit family.
Keywords
3D-structure
4Fe-4S
Acetylation
Complete proteome
Direct protein sequencing
Electron transport
Flavoprotein
FMN
Iron
Iron-sulfur
Membrane
Metal-binding
Methylation
Mitochondrion
Mitochondrion inner membrane
NAD
Oxidoreductase
Reference proteome
Respiratory chain
Transit peptide
Translocase
Transport
Ubiquinone
Feature
chain NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
Uniprot
H9J921
A0A194R3P0
A0A194PZ05
A0A212EVS8
A0A2A4JP06
A0A2H1VYJ4
+ More
A0A182F331 T1DN18 A0A2P8YPW5 A0A2M4BLF0 A0A1B6CWN4 A0A2M4A273 W5JVR1 A0A1W4UXW4 A0A2J7RAB5 A0A0K8TSY0 A0A2M3ZG33 A0A2M3Z095 V5GR67 D6WKB4 A0A182PDX6 A0A182JNE4 A0A182XHC5 A0A182UQQ2 A0A1S4H2T5 A0A182L7X1 A0A182HR46 A0A182R1T4 A0A1A9YLI0 A0A1B6GR77 D3TS33 J3JUS2 A0A067RGT2 A0A182ULL1 A0A182W8L8 A0A182Y655 Q9VMI3 Q7PMH3 A0A1B6GZ57 A0A2S2R2E1 A0A0J9QXJ4 A0A1B0BQ20 A0A1A9ZWA1 J9JIY5 A0A2H8TE50 B4NZQ8 B3MKI8 H9J919 B0WGB3 A0A0M4E3M9 A0A1I8PBI8 B3N9R3 A0A182LXF2 A0A1A9V1J3 T1P7Z7 A0A1A9W856 A0A2K5QWR4 E2R514 A0A182NNV7 U3BG69 A0A1L8EEP0 A0A182QA74 A0A034WMT4 A0A2K5DQ22 A0A2U3YIV9 M3VX83 B4LRL7 A0A034WIS7 B4KL74 A0A2Y9GU77 U3DH76 M3XY56 A0A383Z7F0 B4MUX4 A0A1A6H3E3 F1RVN1 A0A1S2ZFY1 A0A2U3VGJ4 A0A2F0AUY4 Q91YT0 A0A1S3FKS0 A0A1Y1M4Z9 A0A384DJY0 A0A2Y9JN07 A0A3P4QN48 G1L2L0 A0A1U7T3J7 A0A2Y9PMH0 A0A250YIQ0 A0A2Y9DZW7 A0A1B0FE00 A0A2K6A345 A0A2K5L8V7 W5PUX0 A0A1S2ZFW6 B4JB10 A0A2K6DJ47 A0A2K6M2W1 A0A0D9R8L3 A0A2K6QG60
A0A182F331 T1DN18 A0A2P8YPW5 A0A2M4BLF0 A0A1B6CWN4 A0A2M4A273 W5JVR1 A0A1W4UXW4 A0A2J7RAB5 A0A0K8TSY0 A0A2M3ZG33 A0A2M3Z095 V5GR67 D6WKB4 A0A182PDX6 A0A182JNE4 A0A182XHC5 A0A182UQQ2 A0A1S4H2T5 A0A182L7X1 A0A182HR46 A0A182R1T4 A0A1A9YLI0 A0A1B6GR77 D3TS33 J3JUS2 A0A067RGT2 A0A182ULL1 A0A182W8L8 A0A182Y655 Q9VMI3 Q7PMH3 A0A1B6GZ57 A0A2S2R2E1 A0A0J9QXJ4 A0A1B0BQ20 A0A1A9ZWA1 J9JIY5 A0A2H8TE50 B4NZQ8 B3MKI8 H9J919 B0WGB3 A0A0M4E3M9 A0A1I8PBI8 B3N9R3 A0A182LXF2 A0A1A9V1J3 T1P7Z7 A0A1A9W856 A0A2K5QWR4 E2R514 A0A182NNV7 U3BG69 A0A1L8EEP0 A0A182QA74 A0A034WMT4 A0A2K5DQ22 A0A2U3YIV9 M3VX83 B4LRL7 A0A034WIS7 B4KL74 A0A2Y9GU77 U3DH76 M3XY56 A0A383Z7F0 B4MUX4 A0A1A6H3E3 F1RVN1 A0A1S2ZFY1 A0A2U3VGJ4 A0A2F0AUY4 Q91YT0 A0A1S3FKS0 A0A1Y1M4Z9 A0A384DJY0 A0A2Y9JN07 A0A3P4QN48 G1L2L0 A0A1U7T3J7 A0A2Y9PMH0 A0A250YIQ0 A0A2Y9DZW7 A0A1B0FE00 A0A2K6A345 A0A2K5L8V7 W5PUX0 A0A1S2ZFW6 B4JB10 A0A2K6DJ47 A0A2K6M2W1 A0A0D9R8L3 A0A2K6QG60
EC Number
1.6.99.3
7.1.1.2
7.1.1.2
Pubmed
19121390
26354079
22118469
29403074
20920257
23761445
+ More
26369729 18362917 19820115 12364791 20966253 20353571 22516182 23537049 24845553 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17994087 17550304 25315136 16341006 25243066 25348373 17975172 27912063 27654917 16141072 15489334 10870971 21183079 23806337 23576753 24129315 28004739 24813606 20010809 28087693 20809919 27595392 25362486
26369729 18362917 19820115 12364791 20966253 20353571 22516182 23537049 24845553 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17994087 17550304 25315136 16341006 25243066 25348373 17975172 27912063 27654917 16141072 15489334 10870971 21183079 23806337 23576753 24129315 28004739 24813606 20010809 28087693 20809919 27595392 25362486
EMBL
BABH01004638
BABH01004639
BABH01004640
BABH01004641
KQ460855
KPJ11855.1
+ More
KQ459584 KPI98562.1 AGBW02012119 OWR45608.1 NWSH01000867 PCG73795.1 ODYU01005241 SOQ45905.1 GAMD01003250 JAA98340.1 PYGN01000442 PSN46298.1 GGFJ01004746 MBW53887.1 GEDC01019494 JAS17804.1 GGFK01001427 MBW34748.1 ADMH02000277 ETN67150.1 NEVH01006574 PNF37779.1 GDAI01000136 JAI17467.1 GGFM01006649 MBW27400.1 GGFM01001194 MBW21945.1 GALX01004379 JAB64087.1 KQ971342 EFA04664.1 AAAB01008980 APCN01001643 GECZ01004827 JAS64942.1 EZ424235 ADD20511.1 APGK01053537 BT126987 KB741224 KB631749 AEE61949.1 ENN72318.1 ERL85747.1 KK852692 KDR18342.1 AE014134 AY051642 AAF52334.1 AAK93066.1 AHN54172.1 EAA13847.5 GECZ01002075 JAS67694.1 GGMS01014900 MBY84103.1 CM002910 KMY88454.1 JXJN01018356 ABLF02030142 GFXV01000571 MBW12376.1 CM000157 EDW88842.1 CH902620 EDV31541.1 BABH01004644 BABH01004645 DS231924 EDS26851.1 CP012523 ALC38258.1 CH954177 EDV58558.1 AXCM01000658 KA644682 AFP59311.1 AAEX03011611 GAMT01000068 GAMS01002084 GAMP01005499 JAB11793.1 JAB21052.1 JAB47256.1 GFDG01001628 JAV17171.1 AXCN02001989 GAKP01003467 JAC55485.1 AANG04000779 CH940649 EDW63612.1 GAKP01003466 JAC55486.1 CH933807 EDW11735.1 GAMR01002340 JAB31592.1 AEYP01049320 CH963857 EDW76319.1 LZPO01055072 OBS72871.1 AEMK02000006 NTJE010004745 MBV94564.1 AK075692 BC014818 BC041682 GEZM01043063 JAV79345.1 CYRY02037493 VCX24049.1 ACTA01122041 GFFW01001362 JAV43426.1 CCAG010016923 AMGL01061994 CH916368 EDW02880.1 AQIB01078333
KQ459584 KPI98562.1 AGBW02012119 OWR45608.1 NWSH01000867 PCG73795.1 ODYU01005241 SOQ45905.1 GAMD01003250 JAA98340.1 PYGN01000442 PSN46298.1 GGFJ01004746 MBW53887.1 GEDC01019494 JAS17804.1 GGFK01001427 MBW34748.1 ADMH02000277 ETN67150.1 NEVH01006574 PNF37779.1 GDAI01000136 JAI17467.1 GGFM01006649 MBW27400.1 GGFM01001194 MBW21945.1 GALX01004379 JAB64087.1 KQ971342 EFA04664.1 AAAB01008980 APCN01001643 GECZ01004827 JAS64942.1 EZ424235 ADD20511.1 APGK01053537 BT126987 KB741224 KB631749 AEE61949.1 ENN72318.1 ERL85747.1 KK852692 KDR18342.1 AE014134 AY051642 AAF52334.1 AAK93066.1 AHN54172.1 EAA13847.5 GECZ01002075 JAS67694.1 GGMS01014900 MBY84103.1 CM002910 KMY88454.1 JXJN01018356 ABLF02030142 GFXV01000571 MBW12376.1 CM000157 EDW88842.1 CH902620 EDV31541.1 BABH01004644 BABH01004645 DS231924 EDS26851.1 CP012523 ALC38258.1 CH954177 EDV58558.1 AXCM01000658 KA644682 AFP59311.1 AAEX03011611 GAMT01000068 GAMS01002084 GAMP01005499 JAB11793.1 JAB21052.1 JAB47256.1 GFDG01001628 JAV17171.1 AXCN02001989 GAKP01003467 JAC55485.1 AANG04000779 CH940649 EDW63612.1 GAKP01003466 JAC55486.1 CH933807 EDW11735.1 GAMR01002340 JAB31592.1 AEYP01049320 CH963857 EDW76319.1 LZPO01055072 OBS72871.1 AEMK02000006 NTJE010004745 MBV94564.1 AK075692 BC014818 BC041682 GEZM01043063 JAV79345.1 CYRY02037493 VCX24049.1 ACTA01122041 GFFW01001362 JAV43426.1 CCAG010016923 AMGL01061994 CH916368 EDW02880.1 AQIB01078333
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000069272
+ More
UP000245037 UP000000673 UP000192221 UP000235965 UP000007266 UP000075885 UP000075881 UP000076407 UP000075903 UP000075882 UP000075840 UP000075900 UP000092443 UP000019118 UP000030742 UP000027135 UP000075902 UP000075920 UP000076408 UP000000803 UP000007062 UP000092460 UP000092445 UP000007819 UP000002282 UP000007801 UP000002320 UP000092553 UP000095300 UP000008711 UP000075883 UP000078200 UP000095301 UP000091820 UP000233040 UP000002254 UP000075884 UP000008225 UP000075886 UP000233020 UP000245341 UP000011712 UP000008792 UP000009192 UP000248481 UP000000715 UP000261681 UP000007798 UP000092124 UP000008227 UP000079721 UP000245340 UP000000589 UP000081671 UP000261680 UP000291021 UP000248482 UP000008912 UP000189704 UP000248483 UP000248480 UP000092444 UP000233140 UP000233060 UP000002356 UP000001070 UP000233120 UP000233180 UP000029965 UP000233200
UP000245037 UP000000673 UP000192221 UP000235965 UP000007266 UP000075885 UP000075881 UP000076407 UP000075903 UP000075882 UP000075840 UP000075900 UP000092443 UP000019118 UP000030742 UP000027135 UP000075902 UP000075920 UP000076408 UP000000803 UP000007062 UP000092460 UP000092445 UP000007819 UP000002282 UP000007801 UP000002320 UP000092553 UP000095300 UP000008711 UP000075883 UP000078200 UP000095301 UP000091820 UP000233040 UP000002254 UP000075884 UP000008225 UP000075886 UP000233020 UP000245341 UP000011712 UP000008792 UP000009192 UP000248481 UP000000715 UP000261681 UP000007798 UP000092124 UP000008227 UP000079721 UP000245340 UP000000589 UP000081671 UP000261680 UP000291021 UP000248482 UP000008912 UP000189704 UP000248483 UP000248480 UP000092444 UP000233140 UP000233060 UP000002356 UP000001070 UP000233120 UP000233180 UP000029965 UP000233200
Pfam
Interpro
IPR037225
Nuo51_FMN-bd_sf
+ More
IPR019554 Soluble_ligand-bd
IPR011537 NADH-UbQ_OxRdtase_suF
IPR011538 Nuo51_FMN-bd
IPR019575 Nuop51_4Fe4S-bd
IPR001949 NADH-UbQ_OxRdtase_51kDa_CS
IPR037207 Nuop51_4Fe4S-bd_sf
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR029021 Prot-tyrosine_phosphatase-like
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR001339 mRNA_cap_enzyme
IPR000340 Dual-sp_phosphatase_cat-dom
IPR019554 Soluble_ligand-bd
IPR011537 NADH-UbQ_OxRdtase_suF
IPR011538 Nuo51_FMN-bd
IPR019575 Nuop51_4Fe4S-bd
IPR001949 NADH-UbQ_OxRdtase_51kDa_CS
IPR037207 Nuop51_4Fe4S-bd_sf
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR029021 Prot-tyrosine_phosphatase-like
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR001339 mRNA_cap_enzyme
IPR000340 Dual-sp_phosphatase_cat-dom
Gene 3D
ProteinModelPortal
H9J921
A0A194R3P0
A0A194PZ05
A0A212EVS8
A0A2A4JP06
A0A2H1VYJ4
+ More
A0A182F331 T1DN18 A0A2P8YPW5 A0A2M4BLF0 A0A1B6CWN4 A0A2M4A273 W5JVR1 A0A1W4UXW4 A0A2J7RAB5 A0A0K8TSY0 A0A2M3ZG33 A0A2M3Z095 V5GR67 D6WKB4 A0A182PDX6 A0A182JNE4 A0A182XHC5 A0A182UQQ2 A0A1S4H2T5 A0A182L7X1 A0A182HR46 A0A182R1T4 A0A1A9YLI0 A0A1B6GR77 D3TS33 J3JUS2 A0A067RGT2 A0A182ULL1 A0A182W8L8 A0A182Y655 Q9VMI3 Q7PMH3 A0A1B6GZ57 A0A2S2R2E1 A0A0J9QXJ4 A0A1B0BQ20 A0A1A9ZWA1 J9JIY5 A0A2H8TE50 B4NZQ8 B3MKI8 H9J919 B0WGB3 A0A0M4E3M9 A0A1I8PBI8 B3N9R3 A0A182LXF2 A0A1A9V1J3 T1P7Z7 A0A1A9W856 A0A2K5QWR4 E2R514 A0A182NNV7 U3BG69 A0A1L8EEP0 A0A182QA74 A0A034WMT4 A0A2K5DQ22 A0A2U3YIV9 M3VX83 B4LRL7 A0A034WIS7 B4KL74 A0A2Y9GU77 U3DH76 M3XY56 A0A383Z7F0 B4MUX4 A0A1A6H3E3 F1RVN1 A0A1S2ZFY1 A0A2U3VGJ4 A0A2F0AUY4 Q91YT0 A0A1S3FKS0 A0A1Y1M4Z9 A0A384DJY0 A0A2Y9JN07 A0A3P4QN48 G1L2L0 A0A1U7T3J7 A0A2Y9PMH0 A0A250YIQ0 A0A2Y9DZW7 A0A1B0FE00 A0A2K6A345 A0A2K5L8V7 W5PUX0 A0A1S2ZFW6 B4JB10 A0A2K6DJ47 A0A2K6M2W1 A0A0D9R8L3 A0A2K6QG60
A0A182F331 T1DN18 A0A2P8YPW5 A0A2M4BLF0 A0A1B6CWN4 A0A2M4A273 W5JVR1 A0A1W4UXW4 A0A2J7RAB5 A0A0K8TSY0 A0A2M3ZG33 A0A2M3Z095 V5GR67 D6WKB4 A0A182PDX6 A0A182JNE4 A0A182XHC5 A0A182UQQ2 A0A1S4H2T5 A0A182L7X1 A0A182HR46 A0A182R1T4 A0A1A9YLI0 A0A1B6GR77 D3TS33 J3JUS2 A0A067RGT2 A0A182ULL1 A0A182W8L8 A0A182Y655 Q9VMI3 Q7PMH3 A0A1B6GZ57 A0A2S2R2E1 A0A0J9QXJ4 A0A1B0BQ20 A0A1A9ZWA1 J9JIY5 A0A2H8TE50 B4NZQ8 B3MKI8 H9J919 B0WGB3 A0A0M4E3M9 A0A1I8PBI8 B3N9R3 A0A182LXF2 A0A1A9V1J3 T1P7Z7 A0A1A9W856 A0A2K5QWR4 E2R514 A0A182NNV7 U3BG69 A0A1L8EEP0 A0A182QA74 A0A034WMT4 A0A2K5DQ22 A0A2U3YIV9 M3VX83 B4LRL7 A0A034WIS7 B4KL74 A0A2Y9GU77 U3DH76 M3XY56 A0A383Z7F0 B4MUX4 A0A1A6H3E3 F1RVN1 A0A1S2ZFY1 A0A2U3VGJ4 A0A2F0AUY4 Q91YT0 A0A1S3FKS0 A0A1Y1M4Z9 A0A384DJY0 A0A2Y9JN07 A0A3P4QN48 G1L2L0 A0A1U7T3J7 A0A2Y9PMH0 A0A250YIQ0 A0A2Y9DZW7 A0A1B0FE00 A0A2K6A345 A0A2K5L8V7 W5PUX0 A0A1S2ZFW6 B4JB10 A0A2K6DJ47 A0A2K6M2W1 A0A0D9R8L3 A0A2K6QG60
PDB
6G72
E-value=0,
Score=2104
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
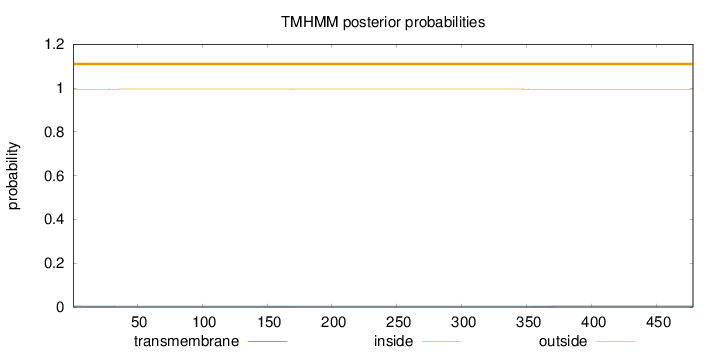
Length:
478
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05829
Exp number, first 60 AAs:
0.00724
Total prob of N-in:
0.00518
outside
1 - 478
Population Genetic Test Statistics
Pi
175.973597
Theta
155.151778
Tajima's D
0.385289
CLR
0.637044
CSRT
0.47777611119444
Interpretation
Uncertain
