Gene
KWMTBOMO01751
Pre Gene Modal
BGIBMGA006136
Annotation
PREDICTED:_tribbles_homolog_2-like_[Papilio_machaon]
Full name
Tribbles homolog 2
Location in the cell
Mitochondrial Reliability : 1.512 Nuclear Reliability : 1.928
Sequence
CDS
ATGGATCCCGGATCACCCTCGTTGTTATGGCCGTTGCGGCTGAGCAAGCCGGCGGTGACGATGCCGGTGCCCAGTACAGGAGAGAGCAGTAAAGCTGAAGACCGAGCGCAGTCCCCAGGGCTGCAGGAGCGGCTGTCCGCGGCGCCACCGAGCCCGGCGCCACCCAGTCCCGCACCAACCCAACATCAATTCCAAGCAGCTCTCGTTGCCGAGAAGTACCTCCTGCTGGAACAAGTCGAAGGTTCCTCGTTATGTCGGTGTGTAGACGTGCGTACCCAAGAGGAATATGTATGCAAGGTTGTGTCCCGAGACTGCAGCAGTTTACTACAAGCACATTACCGTCTCGACGGGCATCCGCACGTCAATCCTATTCACGAAGTCCTAGTCGGCGATAAGAGGGTGTACCTGATCTTCCCTCGGTCGCATTCAGATCTACATTCATACGTTCGGGCGAGGAAAAGGCTAAGAGAGCACGAAGCCCGAAGACTCTTCAGACAAATGGCCGAAACGGTAGCGGCGTGCCACGAACAGGGCATCGTATTAAGAGATCTCAAATTAAGGAAGTTCGTCTTCGCAGACCCACAACGGACGACTCTAAAATTAGAATCATTAGAAGACGCGGTCGTCCTCGACGATCCGGAGGAAGATTTACTTCAAGATAAGCGAGGGTGTCCCGCCTACGTGTCGCCCGAGATCCTCAAAGCGCACAGAACGTATTCGGGCAAGGCCGCCGATATGTGGTCGCTCGGCGTCATACTCTACACCATGCTCGTCGGAAGGTATCCGTTCAACGACTCCGAGCACGCGTCGCTGTTCGCGAAGATATCGCGGGGCTTCTTCGTGGTGCCGGAGTGTTTGTCGTCGCGCGGCAAGTGCCTGATCAGGTCGTTGCTCCGTCGGGAGGCGGGCGAGCGGCTGAGCGCGGCGGCGGTGCTGCGACACCCCTGGCTGGCGCGGGACCCCCGCAACGAGCTGCCGCCGCGCGCCGCCGCCGCGCTCGACCGCCTCGTGCCCGACTGTCTCTAG
Protein
MDPGSPSLLWPLRLSKPAVTMPVPSTGESSKAEDRAQSPGLQERLSAAPPSPAPPSPAPTQHQFQAALVAEKYLLLEQVEGSSLCRCVDVRTQEEYVCKVVSRDCSSLLQAHYRLDGHPHVNPIHEVLVGDKRVYLIFPRSHSDLHSYVRARKRLREHEARRLFRQMAETVAACHEQGIVLRDLKLRKFVFADPQRTTLKLESLEDAVVLDDPEEDLLQDKRGCPAYVSPEILKAHRTYSGKAADMWSLGVILYTMLVGRYPFNDSEHASLFAKISRGFFVVPECLSSRGKCLIRSLLRREAGERLSAAAVLRHPWLARDPRNELPPRAAAALDRLVPDCL
Summary
Description
Interacts with MAPK kinases and regulates activation of MAP kinases. Does not display kinase activity (By similarity).
Similarity
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. Tribbles subfamily.
Keywords
Complete proteome
Cytoplasm
Cytoskeleton
Protein kinase inhibitor
Reference proteome
Feature
chain Tribbles homolog 2
Uniprot
A0A2A4JQB8
A0A1Y1KLV6
A0A1B6DSY4
A0A139WI05
V5GQU8
A0A1B6H5K9
+ More
A0A023EUZ0 A0A1S4G7P6 A0A1B6GLZ5 A0A1Q3FXU2 A0A1Q3FXN3 A0A023EZF0 A0A069DSH6 A0A1L8DKA6 A0A1L8DK86 A0A224XFT1 A0A0N7Z8H9 A0A0P6GQ41 A0A0A9X162 A0A1I8JT69 A0A0V0GF27 A0A2M4AGK8 A0A2M4ABC3 A0A2M3ZGR9 A0A2M4BPI1 A0A2M4BR55 A0A2M4CGS0 A0A2M4A6S5 A0A182Y655 A0A2M4A656 A0A2M4BLU9 A0A2M4CGT5 A0A2M4CGQ3 A0A2M4CHY5 A0A2M4BGP3 A0A0N8EQC5 A0A336LPA0 A0A293MX02 A0A1Z5KXK8 A0A2R5LAD9 B7PAN3 V5HDJ0 A0A1E1XKY1 A0A087UE82 A0A1E1XAM1 G3MIY1 A0A131YPY1 A0A224Z488 A0A131XC18 L7M1C6 L7LRV4 A0A1W7R9C6 A0A154NYQ2 A0A2A3EGS1 A0A195E5M7 A0A151IKT0 A0A310S6C1 A0A158NG97 A0A0C9Q9L5 A0A0M9A3K2 F4WP51 A0A026WQ26 A0A0J7L1E3 E2A3K4 A0A210Q7I2 A0A0L7RJZ6 K7INF4 T1JD19 C3YMQ1 A0A151WMA4 A0A3M6U337 V9K983 A0A091E7V4 A0A2B4S343 A0A1S3EXK5 W5NI83 A0A1S3MBQ9 A0A286XZB7 A0A060XNY0 A0A250YFY7 A7S0A1 G5AST4 A0A1U7R0L9 A0A287BA06 I3M0W8 H0WKU6 H3AA74 Q8K4K3 A0A2I2V3C7 L8I375 Q5GLH2
A0A023EUZ0 A0A1S4G7P6 A0A1B6GLZ5 A0A1Q3FXU2 A0A1Q3FXN3 A0A023EZF0 A0A069DSH6 A0A1L8DKA6 A0A1L8DK86 A0A224XFT1 A0A0N7Z8H9 A0A0P6GQ41 A0A0A9X162 A0A1I8JT69 A0A0V0GF27 A0A2M4AGK8 A0A2M4ABC3 A0A2M3ZGR9 A0A2M4BPI1 A0A2M4BR55 A0A2M4CGS0 A0A2M4A6S5 A0A182Y655 A0A2M4A656 A0A2M4BLU9 A0A2M4CGT5 A0A2M4CGQ3 A0A2M4CHY5 A0A2M4BGP3 A0A0N8EQC5 A0A336LPA0 A0A293MX02 A0A1Z5KXK8 A0A2R5LAD9 B7PAN3 V5HDJ0 A0A1E1XKY1 A0A087UE82 A0A1E1XAM1 G3MIY1 A0A131YPY1 A0A224Z488 A0A131XC18 L7M1C6 L7LRV4 A0A1W7R9C6 A0A154NYQ2 A0A2A3EGS1 A0A195E5M7 A0A151IKT0 A0A310S6C1 A0A158NG97 A0A0C9Q9L5 A0A0M9A3K2 F4WP51 A0A026WQ26 A0A0J7L1E3 E2A3K4 A0A210Q7I2 A0A0L7RJZ6 K7INF4 T1JD19 C3YMQ1 A0A151WMA4 A0A3M6U337 V9K983 A0A091E7V4 A0A2B4S343 A0A1S3EXK5 W5NI83 A0A1S3MBQ9 A0A286XZB7 A0A060XNY0 A0A250YFY7 A7S0A1 G5AST4 A0A1U7R0L9 A0A287BA06 I3M0W8 H0WKU6 H3AA74 Q8K4K3 A0A2I2V3C7 L8I375 Q5GLH2
Pubmed
28004739
18362917
19820115
24945155
25474469
26334808
+ More
27129103 25401762 26823975 25244985 28528879 25765539 29209593 28503490 22216098 26830274 28797301 28049606 25576852 21347285 21719571 24508170 30249741 20798317 28812685 20075255 18563158 30382153 24402279 21993624 24755649 28087693 17615350 21993625 30723633 9215903 16141072 15489334 17975172 22751099 15829623
27129103 25401762 26823975 25244985 28528879 25765539 29209593 28503490 22216098 26830274 28797301 28049606 25576852 21347285 21719571 24508170 30249741 20798317 28812685 20075255 18563158 30382153 24402279 21993624 24755649 28087693 17615350 21993625 30723633 9215903 16141072 15489334 17975172 22751099 15829623
EMBL
NWSH01000867
PCG73794.1
GEZM01086038
JAV59827.1
GEDC01008533
JAS28765.1
+ More
KQ971342 KYB27630.1 GALX01004514 JAB63952.1 GECU01037717 JAS69989.1 GAPW01001299 JAC12299.1 GECZ01006304 JAS63465.1 GFDL01002664 JAV32381.1 GFDL01002704 JAV32341.1 GBBI01004566 JAC14146.1 GBGD01002262 JAC86627.1 GFDF01007195 JAV06889.1 GFDF01007196 JAV06888.1 GFTR01005090 JAW11336.1 GDKW01003299 JAI53296.1 GDIQ01040653 JAN54084.1 GBHO01032764 GBHO01020613 GBRD01010322 GDHC01020797 GDHC01018650 GDHC01018486 JAG10840.1 JAG22991.1 JAG55502.1 JAP97831.1 JAP99978.1 JAQ00143.1 APCN01001643 GECL01000221 JAP05903.1 GGFK01006447 MBW39768.1 GGFK01004770 MBW38091.1 GGFM01006958 MBW27709.1 GGFJ01005773 MBW54914.1 GGFJ01006322 MBW55463.1 GGFL01000358 MBW64536.1 GGFK01003176 MBW36497.1 GGFK01002881 MBW36202.1 GGFJ01004915 MBW54056.1 GGFL01000355 MBW64533.1 GGFL01000354 MBW64532.1 GGFL01000350 MBW64528.1 GGFJ01003084 MBW52225.1 GDIQ01004268 JAN90469.1 UFQT01000030 SSX18197.1 GFWV01019152 MAA43880.1 GFJQ02007097 JAV99872.1 GGLE01002314 MBY06440.1 ABJB010057699 ABJB010608531 DS672165 EEC03655.1 GANP01011241 JAB73227.1 GFAA01003432 JAU00003.1 KK119418 KFM75671.1 GFAC01002888 JAT96300.1 JO841832 AEO33449.1 GEDV01007965 JAP80592.1 GFPF01012849 MAA23995.1 GEFH01004082 JAP64499.1 GACK01008091 JAA56943.1 GACK01010293 JAA54741.1 GFAH01000641 JAV47748.1 KQ434777 KZC04204.1 KZ288253 PBC30965.1 KQ979608 KYN20391.1 KQ977164 KYN05212.1 KQ797156 OAD46995.1 ADTU01015083 ADTU01015084 GBYB01011068 JAG80835.1 KQ435771 KOX75149.1 GL888243 EGI63982.1 KK107139 QOIP01000003 EZA57796.1 RLU24700.1 LBMM01001289 KMQ96572.1 GL436444 EFN71986.1 NEDP02004701 OWF44684.1 KQ414578 KOC71154.1 JH432094 GG666531 EEN58392.1 KQ982944 KYQ48931.1 RCHS01002311 RMX48095.1 JW862171 AFO94688.1 KN122310 KFO31191.1 LSMT01000225 PFX22905.1 AHAT01009048 AAKN02053135 FR905679 CDQ80932.1 GFFW01002245 JAV42543.1 DS469560 EDO42841.1 JH166813 GEBF01005553 EHB00096.1 JAN98079.1 AEMK02000021 DQIR01239618 HDB95095.1 AGTP01047896 AGTP01047897 AAQR03048269 AFYH01219898 AFYH01219899 AFYH01219900 AF358867 AK044747 AK080064 AK082329 AK144192 BC027159 BC034338 BC037387 AAH27159.1 AAH34338.1 AAH37387.1 AAM45477.1 BAC32063.1 BAC37820.1 BAC38467.1 BAE25758.1 AANG04003329 JH882087 ELR50955.1 AY360147 AY247741 BC123595 AAP04410.1 AAR12274.1
KQ971342 KYB27630.1 GALX01004514 JAB63952.1 GECU01037717 JAS69989.1 GAPW01001299 JAC12299.1 GECZ01006304 JAS63465.1 GFDL01002664 JAV32381.1 GFDL01002704 JAV32341.1 GBBI01004566 JAC14146.1 GBGD01002262 JAC86627.1 GFDF01007195 JAV06889.1 GFDF01007196 JAV06888.1 GFTR01005090 JAW11336.1 GDKW01003299 JAI53296.1 GDIQ01040653 JAN54084.1 GBHO01032764 GBHO01020613 GBRD01010322 GDHC01020797 GDHC01018650 GDHC01018486 JAG10840.1 JAG22991.1 JAG55502.1 JAP97831.1 JAP99978.1 JAQ00143.1 APCN01001643 GECL01000221 JAP05903.1 GGFK01006447 MBW39768.1 GGFK01004770 MBW38091.1 GGFM01006958 MBW27709.1 GGFJ01005773 MBW54914.1 GGFJ01006322 MBW55463.1 GGFL01000358 MBW64536.1 GGFK01003176 MBW36497.1 GGFK01002881 MBW36202.1 GGFJ01004915 MBW54056.1 GGFL01000355 MBW64533.1 GGFL01000354 MBW64532.1 GGFL01000350 MBW64528.1 GGFJ01003084 MBW52225.1 GDIQ01004268 JAN90469.1 UFQT01000030 SSX18197.1 GFWV01019152 MAA43880.1 GFJQ02007097 JAV99872.1 GGLE01002314 MBY06440.1 ABJB010057699 ABJB010608531 DS672165 EEC03655.1 GANP01011241 JAB73227.1 GFAA01003432 JAU00003.1 KK119418 KFM75671.1 GFAC01002888 JAT96300.1 JO841832 AEO33449.1 GEDV01007965 JAP80592.1 GFPF01012849 MAA23995.1 GEFH01004082 JAP64499.1 GACK01008091 JAA56943.1 GACK01010293 JAA54741.1 GFAH01000641 JAV47748.1 KQ434777 KZC04204.1 KZ288253 PBC30965.1 KQ979608 KYN20391.1 KQ977164 KYN05212.1 KQ797156 OAD46995.1 ADTU01015083 ADTU01015084 GBYB01011068 JAG80835.1 KQ435771 KOX75149.1 GL888243 EGI63982.1 KK107139 QOIP01000003 EZA57796.1 RLU24700.1 LBMM01001289 KMQ96572.1 GL436444 EFN71986.1 NEDP02004701 OWF44684.1 KQ414578 KOC71154.1 JH432094 GG666531 EEN58392.1 KQ982944 KYQ48931.1 RCHS01002311 RMX48095.1 JW862171 AFO94688.1 KN122310 KFO31191.1 LSMT01000225 PFX22905.1 AHAT01009048 AAKN02053135 FR905679 CDQ80932.1 GFFW01002245 JAV42543.1 DS469560 EDO42841.1 JH166813 GEBF01005553 EHB00096.1 JAN98079.1 AEMK02000021 DQIR01239618 HDB95095.1 AGTP01047896 AGTP01047897 AAQR03048269 AFYH01219898 AFYH01219899 AFYH01219900 AF358867 AK044747 AK080064 AK082329 AK144192 BC027159 BC034338 BC037387 AAH27159.1 AAH34338.1 AAH37387.1 AAM45477.1 BAC32063.1 BAC37820.1 BAC38467.1 BAE25758.1 AANG04003329 JH882087 ELR50955.1 AY360147 AY247741 BC123595 AAP04410.1 AAR12274.1
Proteomes
UP000218220
UP000007266
UP000075840
UP000076408
UP000001555
UP000054359
+ More
UP000076502 UP000242457 UP000078492 UP000078542 UP000005205 UP000053105 UP000007755 UP000053097 UP000279307 UP000036403 UP000000311 UP000242188 UP000053825 UP000002358 UP000001554 UP000075809 UP000275408 UP000028990 UP000225706 UP000081671 UP000018468 UP000087266 UP000005447 UP000193380 UP000001593 UP000006813 UP000189706 UP000008227 UP000005215 UP000005225 UP000008672 UP000000589 UP000011712 UP000009136
UP000076502 UP000242457 UP000078492 UP000078542 UP000005205 UP000053105 UP000007755 UP000053097 UP000279307 UP000036403 UP000000311 UP000242188 UP000053825 UP000002358 UP000001554 UP000075809 UP000275408 UP000028990 UP000225706 UP000081671 UP000018468 UP000087266 UP000005447 UP000193380 UP000001593 UP000006813 UP000189706 UP000008227 UP000005215 UP000005225 UP000008672 UP000000589 UP000011712 UP000009136
Interpro
IPR000719
Prot_kinase_dom
+ More
IPR011009 Kinase-like_dom_sf
IPR024104 Tribbles/Ser_Thr_kinase_40
IPR011538 Nuo51_FMN-bd
IPR011537 NADH-UbQ_OxRdtase_suF
IPR019554 Soluble_ligand-bd
IPR037225 Nuo51_FMN-bd_sf
IPR037207 Nuop51_4Fe4S-bd_sf
IPR001949 NADH-UbQ_OxRdtase_51kDa_CS
IPR019575 Nuop51_4Fe4S-bd
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR024106 Tribbles_TRB3
IPR011009 Kinase-like_dom_sf
IPR024104 Tribbles/Ser_Thr_kinase_40
IPR011538 Nuo51_FMN-bd
IPR011537 NADH-UbQ_OxRdtase_suF
IPR019554 Soluble_ligand-bd
IPR037225 Nuo51_FMN-bd_sf
IPR037207 Nuop51_4Fe4S-bd_sf
IPR001949 NADH-UbQ_OxRdtase_51kDa_CS
IPR019575 Nuop51_4Fe4S-bd
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR024106 Tribbles_TRB3
Gene 3D
ProteinModelPortal
A0A2A4JQB8
A0A1Y1KLV6
A0A1B6DSY4
A0A139WI05
V5GQU8
A0A1B6H5K9
+ More
A0A023EUZ0 A0A1S4G7P6 A0A1B6GLZ5 A0A1Q3FXU2 A0A1Q3FXN3 A0A023EZF0 A0A069DSH6 A0A1L8DKA6 A0A1L8DK86 A0A224XFT1 A0A0N7Z8H9 A0A0P6GQ41 A0A0A9X162 A0A1I8JT69 A0A0V0GF27 A0A2M4AGK8 A0A2M4ABC3 A0A2M3ZGR9 A0A2M4BPI1 A0A2M4BR55 A0A2M4CGS0 A0A2M4A6S5 A0A182Y655 A0A2M4A656 A0A2M4BLU9 A0A2M4CGT5 A0A2M4CGQ3 A0A2M4CHY5 A0A2M4BGP3 A0A0N8EQC5 A0A336LPA0 A0A293MX02 A0A1Z5KXK8 A0A2R5LAD9 B7PAN3 V5HDJ0 A0A1E1XKY1 A0A087UE82 A0A1E1XAM1 G3MIY1 A0A131YPY1 A0A224Z488 A0A131XC18 L7M1C6 L7LRV4 A0A1W7R9C6 A0A154NYQ2 A0A2A3EGS1 A0A195E5M7 A0A151IKT0 A0A310S6C1 A0A158NG97 A0A0C9Q9L5 A0A0M9A3K2 F4WP51 A0A026WQ26 A0A0J7L1E3 E2A3K4 A0A210Q7I2 A0A0L7RJZ6 K7INF4 T1JD19 C3YMQ1 A0A151WMA4 A0A3M6U337 V9K983 A0A091E7V4 A0A2B4S343 A0A1S3EXK5 W5NI83 A0A1S3MBQ9 A0A286XZB7 A0A060XNY0 A0A250YFY7 A7S0A1 G5AST4 A0A1U7R0L9 A0A287BA06 I3M0W8 H0WKU6 H3AA74 Q8K4K3 A0A2I2V3C7 L8I375 Q5GLH2
A0A023EUZ0 A0A1S4G7P6 A0A1B6GLZ5 A0A1Q3FXU2 A0A1Q3FXN3 A0A023EZF0 A0A069DSH6 A0A1L8DKA6 A0A1L8DK86 A0A224XFT1 A0A0N7Z8H9 A0A0P6GQ41 A0A0A9X162 A0A1I8JT69 A0A0V0GF27 A0A2M4AGK8 A0A2M4ABC3 A0A2M3ZGR9 A0A2M4BPI1 A0A2M4BR55 A0A2M4CGS0 A0A2M4A6S5 A0A182Y655 A0A2M4A656 A0A2M4BLU9 A0A2M4CGT5 A0A2M4CGQ3 A0A2M4CHY5 A0A2M4BGP3 A0A0N8EQC5 A0A336LPA0 A0A293MX02 A0A1Z5KXK8 A0A2R5LAD9 B7PAN3 V5HDJ0 A0A1E1XKY1 A0A087UE82 A0A1E1XAM1 G3MIY1 A0A131YPY1 A0A224Z488 A0A131XC18 L7M1C6 L7LRV4 A0A1W7R9C6 A0A154NYQ2 A0A2A3EGS1 A0A195E5M7 A0A151IKT0 A0A310S6C1 A0A158NG97 A0A0C9Q9L5 A0A0M9A3K2 F4WP51 A0A026WQ26 A0A0J7L1E3 E2A3K4 A0A210Q7I2 A0A0L7RJZ6 K7INF4 T1JD19 C3YMQ1 A0A151WMA4 A0A3M6U337 V9K983 A0A091E7V4 A0A2B4S343 A0A1S3EXK5 W5NI83 A0A1S3MBQ9 A0A286XZB7 A0A060XNY0 A0A250YFY7 A7S0A1 G5AST4 A0A1U7R0L9 A0A287BA06 I3M0W8 H0WKU6 H3AA74 Q8K4K3 A0A2I2V3C7 L8I375 Q5GLH2
PDB
5CEM
E-value=6.48383e-68,
Score=653
Ontologies
GO
GO:0005524
GO:0004672
GO:0003714
GO:0031434
GO:0043405
GO:0005634
GO:0032436
GO:0004674
GO:0008137
GO:0051539
GO:0010181
GO:0051287
GO:0004579
GO:0031625
GO:0045599
GO:0055106
GO:0008134
GO:0045081
GO:0001947
GO:0005737
GO:0006468
GO:0004860
GO:0043433
GO:0005856
GO:0051443
GO:0008270
GO:0006396
GO:0030529
GO:0016020
GO:0005515
GO:0003779
GO:0016021
PANTHER
Topology
Subcellular location
Cytoplasm
May associate with the cytoskeleton. With evidence from 1 publications.
Cytoskeleton May associate with the cytoskeleton. With evidence from 1 publications.
Cytoskeleton May associate with the cytoskeleton. With evidence from 1 publications.
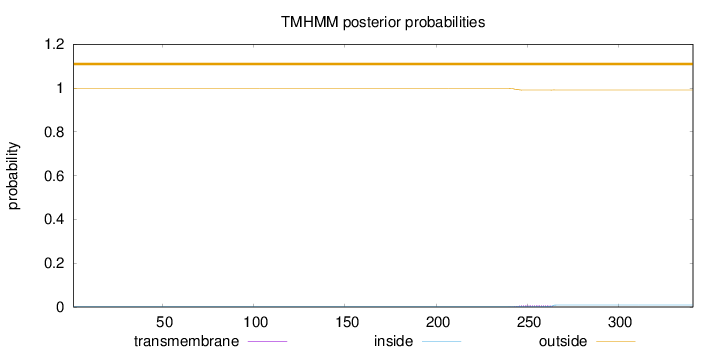
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15612
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00229
outside
1 - 341
Population Genetic Test Statistics
Pi
166.674819
Theta
179.72692
Tajima's D
-0.671636
CLR
148.978192
CSRT
0.206789660516974
Interpretation
Uncertain
