Pre Gene Modal
BGIBMGA006014
Annotation
PREDICTED:_transcription_initiation_factor_IIB_[Amyelois_transitella]
Full name
Transcription initiation factor IIB
Alternative Name
General transcription factor TFIIB
Location in the cell
Nuclear Reliability : 2.115
Sequence
CDS
ATGGCGAGTACATCGAGAATTGAGGCGAATAAAGTGGTTTGCTATGCTCACCCTGATGCGCCCCTTATCGAGGATTACAGAGCGGGTGATATGATATGTTCTGAATGCGGTTTGGTGGTCGGAGACAGAGTCATAGACGTCGGTTCGGAGTGGCGTACTTTTAGCAATGAGAAGTCTGGAGTAGATCCGTCTCGTGTTGGTGGTCCTGAAAACCCACTTTTATCTGGAGGTGATTTGTCAACTATCATTGGGCCAGGACGAGGAGATGCATCATTTGACAGCTTTGGAGTTTCCAAATATCAAAATAGAAGAAACATAAGCAGCACAGATAGAGCCTTAATCAATGCATTCCGAGAAATCAACACAATGGCGGATAGAATTAATCTTCCAAAAACTATTGTGGACAGAGCAAACAATCTTTTTAAACAGGTTCACGACGGTAAAAACTTGAAGGGTCGTGCCAATGATGCTATAGCTTCAGCTTGCCTCTACATTGCTTGTCGTCAAGAAGGTGTGCCACGTACATTTAAAGAGATCTGTGCCGTCAGCAAGATAAGTAAAAAAGAAATAGGAAGGTGTTTCAAATTGATTCTGAAAGCACTGGAGACTTCAGTTGATCTCATTACAACAGCAGATTTTATGTCTCGGTTTTGCTCGAATTTGGGTCTGCCAAATTCAGTGCAACGCGCAGCTACACATATCGCAAGGAAAGCTGGAGAGCTGGACATAGTGTCTGGAAGAAGTCCTATTTCAGTTGCTGCAGCTGCCATTTATATGGCTTCACAGGCATCAGAAGACAAACGCAGTCAGAAAGAGATCGGCGATATTGCTGGTGTTGCGGACGTTACGATCCGACAGTCTTACAAATTGATGTATCCATCCGCAGCAAGACTGTTCCCTGAAGATTTCAAATTCGCAACTCCTATTGAATTCCTACCACAGATGTAG
Protein
MASTSRIEANKVVCYAHPDAPLIEDYRAGDMICSECGLVVGDRVIDVGSEWRTFSNEKSGVDPSRVGGPENPLLSGGDLSTIIGPGRGDASFDSFGVSKYQNRRNISSTDRALINAFREINTMADRINLPKTIVDRANNLFKQVHDGKNLKGRANDAIASACLYIACRQEGVPRTFKEICAVSKISKKEIGRCFKLILKALETSVDLITTADFMSRFCSNLGLPNSVQRAATHIARKAGELDIVSGRSPISVAAAAIYMASQASEDKRSQKEIGDIAGVADVTIRQSYKLMYPSAARLFPEDFKFATPIEFLPQM
Summary
Subunit
Belongs to the TFIID complex which is composed of TATA binding protein (Tbp) and a number of TBP-associated factors (Tafs). Associates with TFIID-IIA (DA complex) to form TFIID-IIA-IIB (DAB-complex) which is then recognized by polymerase II.
Similarity
Belongs to the TFIIB family.
Keywords
Complete proteome
Metal-binding
Nucleus
Reference proteome
Repeat
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Transcription initiation factor IIB
Uniprot
H9J922
A0A212FDG8
A0A2A4JX02
A0A194PZU9
A0A2H1W5H6
A0A194R7K3
+ More
A0A0L7QX69 A0A088A0P9 A0A067R015 A0A2J7QYZ6 D6WN74 A0A1B6GJ52 A0A1B6HU03 A0A0N0BL31 A0A1Y1NCQ2 E2AFR1 A0A026WKH8 J3JZI8 A0A195CTR9 A0A154P0R6 A0A1B6MM13 A0A0M5IWI2 A0A2A3EQY6 E2C0J3 B4LVD1 B4KHS4 A0A0J7L5W8 A0A151JQZ6 A0A158NTU6 B4JPZ0 A0A1W4UFZ1 A0A3B0KDE6 B5DHX4 B4G786 M9PCM4 B3MJI1 B4MUC1 A0A0L0C7U4 B3N9Q3 B4Q9A4 P29052 E0VNE5 A0A232FA54 K7IUT4 B4NZP8 A0A1I8M1Y3 A0A224XSR1 B4HWP7 A0A1I8Q423 A0A195BMG1 A0A195FQY2 A0A151WLM7 A0A0P4VST3 A0A0V0G6F3 A0A1L8DH73 B0WI57 E9IQ81 A0A1S3DGN6 A0A1Q3FWI8 A0A023EPE7 A0A336KY44 Q17AB8 A0A1A9UNQ2 U5EXJ6 A0A1A9ZXG2 A0A1A9W0P8 D3TNF6 A0A1A9XTY1 A0A1B0C246 A0A0K8TRN1 Q17AB9 A0A146LAR9 A0A182GZI9 A0A1B6CE83 A0A182RPR5 A0A182IDZ9 A0A182XCM3 A0A182VV34 A0A0K8W6R6 A0A0K8U1S1 A0A034WT36 A0A034WUD8 W8C7B0 A0A182MQF3 Q7QG89 A0A182UP06 A0A182J3Y5 A0A182NLK2 A0A182L5U4 A0A182Q327 A0A182PB41 A0A182Y5Q4 A0A2M3ZA79 A0A2M4AM58 A0A2M4BUK3 W5J5M4 A0A182F210 A0A1J1HJN0 A0A0P6AIJ9 A0A0P5DJT8
A0A0L7QX69 A0A088A0P9 A0A067R015 A0A2J7QYZ6 D6WN74 A0A1B6GJ52 A0A1B6HU03 A0A0N0BL31 A0A1Y1NCQ2 E2AFR1 A0A026WKH8 J3JZI8 A0A195CTR9 A0A154P0R6 A0A1B6MM13 A0A0M5IWI2 A0A2A3EQY6 E2C0J3 B4LVD1 B4KHS4 A0A0J7L5W8 A0A151JQZ6 A0A158NTU6 B4JPZ0 A0A1W4UFZ1 A0A3B0KDE6 B5DHX4 B4G786 M9PCM4 B3MJI1 B4MUC1 A0A0L0C7U4 B3N9Q3 B4Q9A4 P29052 E0VNE5 A0A232FA54 K7IUT4 B4NZP8 A0A1I8M1Y3 A0A224XSR1 B4HWP7 A0A1I8Q423 A0A195BMG1 A0A195FQY2 A0A151WLM7 A0A0P4VST3 A0A0V0G6F3 A0A1L8DH73 B0WI57 E9IQ81 A0A1S3DGN6 A0A1Q3FWI8 A0A023EPE7 A0A336KY44 Q17AB8 A0A1A9UNQ2 U5EXJ6 A0A1A9ZXG2 A0A1A9W0P8 D3TNF6 A0A1A9XTY1 A0A1B0C246 A0A0K8TRN1 Q17AB9 A0A146LAR9 A0A182GZI9 A0A1B6CE83 A0A182RPR5 A0A182IDZ9 A0A182XCM3 A0A182VV34 A0A0K8W6R6 A0A0K8U1S1 A0A034WT36 A0A034WUD8 W8C7B0 A0A182MQF3 Q7QG89 A0A182UP06 A0A182J3Y5 A0A182NLK2 A0A182L5U4 A0A182Q327 A0A182PB41 A0A182Y5Q4 A0A2M3ZA79 A0A2M4AM58 A0A2M4BUK3 W5J5M4 A0A182F210 A0A1J1HJN0 A0A0P6AIJ9 A0A0P5DJT8
Pubmed
19121390
22118469
26354079
24845553
18362917
19820115
+ More
28004739 20798317 24508170 22516182 23537049 17994087 21347285 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 22936249 1557390 1644295 20566863 28648823 20075255 17550304 25315136 27129103 21282665 24945155 17510324 20353571 26369729 26823975 26483478 25348373 24495485 12364791 14747013 17210077 20966253 25244985 20920257 23761445
28004739 20798317 24508170 22516182 23537049 17994087 21347285 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 22936249 1557390 1644295 20566863 28648823 20075255 17550304 25315136 27129103 21282665 24945155 17510324 20353571 26369729 26823975 26483478 25348373 24495485 12364791 14747013 17210077 20966253 25244985 20920257 23761445
EMBL
BABH01004630
AGBW02009044
OWR51796.1
NWSH01000448
PCG76339.1
KQ459584
+ More
KPI98558.1 ODYU01006472 SOQ48339.1 KQ460855 KPJ11851.1 KQ414705 KOC63190.1 KK852840 KDR15192.1 NEVH01009081 PNF33800.1 KQ971342 EFA03052.1 GECZ01007436 JAS62333.1 GECU01029554 JAS78152.1 KQ435687 KOX81398.1 GEZM01007000 JAV95489.1 GL439124 EFN67722.1 KK107167 EZA56488.1 APGK01048267 BT128670 KB741103 KB632401 AEE63627.1 ENN73844.1 ERL94911.1 KQ977279 KYN04086.1 KQ434783 KZC04944.1 GEBQ01002997 JAT36980.1 CP012523 ALC39268.1 KZ288201 PBC33549.1 GL451800 EFN78510.1 CH940649 EDW63310.1 CH933807 EDW13361.1 LBMM01000651 KMQ97889.1 KQ978615 KYN29818.1 ADTU01026159 CH916372 EDV98970.1 OUUW01000006 SPP81678.1 CH379059 EDY69901.2 CH479180 EDW28340.1 AE014134 AGB92884.1 CH902620 EDV32349.1 CH963857 EDW76047.1 JRES01000893 KNC27479.1 CH954177 EDV58548.1 CM000361 CM002910 EDX04546.1 KMY89545.1 M88164 M91081 U02879 U35148 BT011459 BT015269 DS235336 EEB14901.1 NNAY01000550 OXU27726.1 AAZX01002635 CM000157 EDW88832.1 GFTR01004946 JAW11480.1 CH480818 EDW52442.1 KQ976435 KYM87065.1 KQ981382 KYN42314.1 KQ982959 KYQ48764.1 GDKW01001044 JAI55551.1 GECL01002543 JAP03581.1 GFDF01008374 JAV05710.1 DS231943 EDS28248.1 GL764666 EFZ17278.1 GFDL01003137 JAV31908.1 GAPW01002723 JAC10875.1 UFQS01001200 UFQT01001200 SSX09667.1 SSX29457.1 CH477337 EAT43173.1 GANO01000919 JAB58952.1 CCAG010011567 EZ422958 ADD19234.1 JXJN01024340 GDAI01000802 JAI16801.1 EAT43172.1 GDHC01014417 JAQ04212.1 JXUM01099634 KQ564507 KXJ72164.1 GEDC01025587 JAS11711.1 GDHF01005724 JAI46590.1 GDHF01031836 JAI20478.1 GAKP01001148 JAC57804.1 GAKP01001147 JAC57805.1 GAMC01000642 JAC05914.1 AXCM01002280 AAAB01008839 EAA05904.4 AXCN02002502 GGFM01004696 MBW25447.1 GGFK01008539 MBW41860.1 GGFJ01007531 MBW56672.1 ADMH02002162 ETN58170.1 CVRI01000001 CRK86457.1 GDIP01029253 JAM74462.1 GDIP01155226 JAJ68176.1
KPI98558.1 ODYU01006472 SOQ48339.1 KQ460855 KPJ11851.1 KQ414705 KOC63190.1 KK852840 KDR15192.1 NEVH01009081 PNF33800.1 KQ971342 EFA03052.1 GECZ01007436 JAS62333.1 GECU01029554 JAS78152.1 KQ435687 KOX81398.1 GEZM01007000 JAV95489.1 GL439124 EFN67722.1 KK107167 EZA56488.1 APGK01048267 BT128670 KB741103 KB632401 AEE63627.1 ENN73844.1 ERL94911.1 KQ977279 KYN04086.1 KQ434783 KZC04944.1 GEBQ01002997 JAT36980.1 CP012523 ALC39268.1 KZ288201 PBC33549.1 GL451800 EFN78510.1 CH940649 EDW63310.1 CH933807 EDW13361.1 LBMM01000651 KMQ97889.1 KQ978615 KYN29818.1 ADTU01026159 CH916372 EDV98970.1 OUUW01000006 SPP81678.1 CH379059 EDY69901.2 CH479180 EDW28340.1 AE014134 AGB92884.1 CH902620 EDV32349.1 CH963857 EDW76047.1 JRES01000893 KNC27479.1 CH954177 EDV58548.1 CM000361 CM002910 EDX04546.1 KMY89545.1 M88164 M91081 U02879 U35148 BT011459 BT015269 DS235336 EEB14901.1 NNAY01000550 OXU27726.1 AAZX01002635 CM000157 EDW88832.1 GFTR01004946 JAW11480.1 CH480818 EDW52442.1 KQ976435 KYM87065.1 KQ981382 KYN42314.1 KQ982959 KYQ48764.1 GDKW01001044 JAI55551.1 GECL01002543 JAP03581.1 GFDF01008374 JAV05710.1 DS231943 EDS28248.1 GL764666 EFZ17278.1 GFDL01003137 JAV31908.1 GAPW01002723 JAC10875.1 UFQS01001200 UFQT01001200 SSX09667.1 SSX29457.1 CH477337 EAT43173.1 GANO01000919 JAB58952.1 CCAG010011567 EZ422958 ADD19234.1 JXJN01024340 GDAI01000802 JAI16801.1 EAT43172.1 GDHC01014417 JAQ04212.1 JXUM01099634 KQ564507 KXJ72164.1 GEDC01025587 JAS11711.1 GDHF01005724 JAI46590.1 GDHF01031836 JAI20478.1 GAKP01001148 JAC57804.1 GAKP01001147 JAC57805.1 GAMC01000642 JAC05914.1 AXCM01002280 AAAB01008839 EAA05904.4 AXCN02002502 GGFM01004696 MBW25447.1 GGFK01008539 MBW41860.1 GGFJ01007531 MBW56672.1 ADMH02002162 ETN58170.1 CVRI01000001 CRK86457.1 GDIP01029253 JAM74462.1 GDIP01155226 JAJ68176.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000053825
+ More
UP000005203 UP000027135 UP000235965 UP000007266 UP000053105 UP000000311 UP000053097 UP000019118 UP000030742 UP000078542 UP000076502 UP000092553 UP000242457 UP000008237 UP000008792 UP000009192 UP000036403 UP000078492 UP000005205 UP000001070 UP000192221 UP000268350 UP000001819 UP000008744 UP000000803 UP000007801 UP000007798 UP000037069 UP000008711 UP000000304 UP000009046 UP000215335 UP000002358 UP000002282 UP000095301 UP000001292 UP000095300 UP000078540 UP000078541 UP000075809 UP000002320 UP000079169 UP000008820 UP000078200 UP000092445 UP000091820 UP000092444 UP000092443 UP000092460 UP000069940 UP000249989 UP000075900 UP000075920 UP000075883 UP000007062 UP000075903 UP000075880 UP000075884 UP000075882 UP000075886 UP000075885 UP000076408 UP000000673 UP000069272 UP000183832
UP000005203 UP000027135 UP000235965 UP000007266 UP000053105 UP000000311 UP000053097 UP000019118 UP000030742 UP000078542 UP000076502 UP000092553 UP000242457 UP000008237 UP000008792 UP000009192 UP000036403 UP000078492 UP000005205 UP000001070 UP000192221 UP000268350 UP000001819 UP000008744 UP000000803 UP000007801 UP000007798 UP000037069 UP000008711 UP000000304 UP000009046 UP000215335 UP000002358 UP000002282 UP000095301 UP000001292 UP000095300 UP000078540 UP000078541 UP000075809 UP000002320 UP000079169 UP000008820 UP000078200 UP000092445 UP000091820 UP000092444 UP000092443 UP000092460 UP000069940 UP000249989 UP000075900 UP000075920 UP000075883 UP000007062 UP000075903 UP000075880 UP000075884 UP000075882 UP000075886 UP000075885 UP000076408 UP000000673 UP000069272 UP000183832
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J922
A0A212FDG8
A0A2A4JX02
A0A194PZU9
A0A2H1W5H6
A0A194R7K3
+ More
A0A0L7QX69 A0A088A0P9 A0A067R015 A0A2J7QYZ6 D6WN74 A0A1B6GJ52 A0A1B6HU03 A0A0N0BL31 A0A1Y1NCQ2 E2AFR1 A0A026WKH8 J3JZI8 A0A195CTR9 A0A154P0R6 A0A1B6MM13 A0A0M5IWI2 A0A2A3EQY6 E2C0J3 B4LVD1 B4KHS4 A0A0J7L5W8 A0A151JQZ6 A0A158NTU6 B4JPZ0 A0A1W4UFZ1 A0A3B0KDE6 B5DHX4 B4G786 M9PCM4 B3MJI1 B4MUC1 A0A0L0C7U4 B3N9Q3 B4Q9A4 P29052 E0VNE5 A0A232FA54 K7IUT4 B4NZP8 A0A1I8M1Y3 A0A224XSR1 B4HWP7 A0A1I8Q423 A0A195BMG1 A0A195FQY2 A0A151WLM7 A0A0P4VST3 A0A0V0G6F3 A0A1L8DH73 B0WI57 E9IQ81 A0A1S3DGN6 A0A1Q3FWI8 A0A023EPE7 A0A336KY44 Q17AB8 A0A1A9UNQ2 U5EXJ6 A0A1A9ZXG2 A0A1A9W0P8 D3TNF6 A0A1A9XTY1 A0A1B0C246 A0A0K8TRN1 Q17AB9 A0A146LAR9 A0A182GZI9 A0A1B6CE83 A0A182RPR5 A0A182IDZ9 A0A182XCM3 A0A182VV34 A0A0K8W6R6 A0A0K8U1S1 A0A034WT36 A0A034WUD8 W8C7B0 A0A182MQF3 Q7QG89 A0A182UP06 A0A182J3Y5 A0A182NLK2 A0A182L5U4 A0A182Q327 A0A182PB41 A0A182Y5Q4 A0A2M3ZA79 A0A2M4AM58 A0A2M4BUK3 W5J5M4 A0A182F210 A0A1J1HJN0 A0A0P6AIJ9 A0A0P5DJT8
A0A0L7QX69 A0A088A0P9 A0A067R015 A0A2J7QYZ6 D6WN74 A0A1B6GJ52 A0A1B6HU03 A0A0N0BL31 A0A1Y1NCQ2 E2AFR1 A0A026WKH8 J3JZI8 A0A195CTR9 A0A154P0R6 A0A1B6MM13 A0A0M5IWI2 A0A2A3EQY6 E2C0J3 B4LVD1 B4KHS4 A0A0J7L5W8 A0A151JQZ6 A0A158NTU6 B4JPZ0 A0A1W4UFZ1 A0A3B0KDE6 B5DHX4 B4G786 M9PCM4 B3MJI1 B4MUC1 A0A0L0C7U4 B3N9Q3 B4Q9A4 P29052 E0VNE5 A0A232FA54 K7IUT4 B4NZP8 A0A1I8M1Y3 A0A224XSR1 B4HWP7 A0A1I8Q423 A0A195BMG1 A0A195FQY2 A0A151WLM7 A0A0P4VST3 A0A0V0G6F3 A0A1L8DH73 B0WI57 E9IQ81 A0A1S3DGN6 A0A1Q3FWI8 A0A023EPE7 A0A336KY44 Q17AB8 A0A1A9UNQ2 U5EXJ6 A0A1A9ZXG2 A0A1A9W0P8 D3TNF6 A0A1A9XTY1 A0A1B0C246 A0A0K8TRN1 Q17AB9 A0A146LAR9 A0A182GZI9 A0A1B6CE83 A0A182RPR5 A0A182IDZ9 A0A182XCM3 A0A182VV34 A0A0K8W6R6 A0A0K8U1S1 A0A034WT36 A0A034WUD8 W8C7B0 A0A182MQF3 Q7QG89 A0A182UP06 A0A182J3Y5 A0A182NLK2 A0A182L5U4 A0A182Q327 A0A182PB41 A0A182Y5Q4 A0A2M3ZA79 A0A2M4AM58 A0A2M4BUK3 W5J5M4 A0A182F210 A0A1J1HJN0 A0A0P6AIJ9 A0A0P5DJT8
PDB
6O9L
E-value=1.17099e-134,
Score=1228
Ontologies
GO
PANTHER
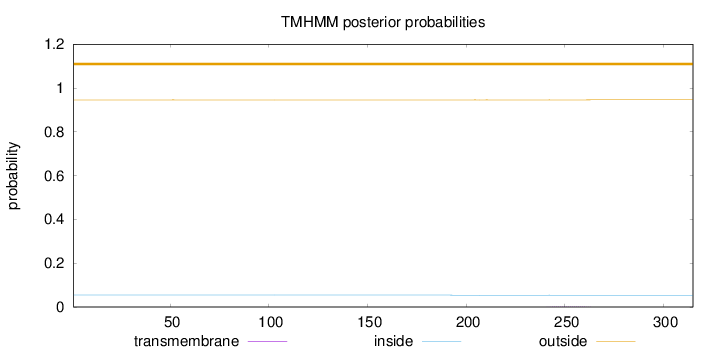
Topology
Subcellular location
Nucleus
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02887
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05427
outside
1 - 315
Population Genetic Test Statistics
Pi
189.121431
Theta
177.147081
Tajima's D
0.229229
CLR
0.745688
CSRT
0.433578321083946
Interpretation
Uncertain
