Gene
KWMTBOMO01745
Annotation
hypothetical_protein_RR46_03708_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.449
Sequence
CDS
ATGTGGTGCACAGGCGTCAATCACTGTAACTTTTTGCTGTCGGAGGATTATCCTCCGTCCAGCAGCTGGTCTCCAGGAGTCGTCTACATCAAATACGCTTGTTTTGATGATACAATATCTCTGCATTACTGCAACCGCGAGTTGAGGATAGCTGAATGGGGAGCCGAGAGCGAGGGTTACATTCGAACCCCAGGGTACCCGCACTTCTACGTTGGCGATGAATGCCGCTGGCGGCTCATTGCCAACCCCGAACAAAGGATACGGGTCACATTCTTGGACGTTAGCTTAAGAAGTATAGGTCCGTTTGAAAACGAGTGTAAGGATTTCGTCTCGGTCCAAGATTCGAATGGCGATAACCTGTTGTACTCGTGCGAACAAGTCGATATGCCGATCAAACTCACTTCCACAACCAACACTGTGGAGGTGGCCGTGGAAGCAAGGTCTAAAGGCGCTTACCCGAAACGAGGAGTCCTGTTGCATTATAAAAGCATAGGCTGTGTTACTTTATCAGCGCCATCTAGTGGATATCTTGTGTATCGCAACGAAGACGTGGCACACTACATGTGCAACGTTAACTTTGTTTTCGTCGACACTCGACAGAGGGCGCGATTACTTTGGTGCTATGACGATAACAGATGGAATAATACCGTGCCTCTCTGCATTGCCGAGACGGTTGTCAAAGCCGTCGGGAACGACTCTTCACGGGAGGGCATCGGCAAAGAAACAAAACTGCCGCCCCACCAAGAACCCAACATGATAGTCGACATCGTCATCCCGTCGCTGCTGATAGCGGCCCTGTTCATCGGAAACGCGTTCATCGTACTCATCATCTATAAATACAGGAAACGGTCAGTTGTTATTGTAGTGTACGGTACTCACATTGTGTGCCCATTTCAAGTACTTCCAAACGTAGATAAGTGA
Protein
MWCTGVNHCNFLLSEDYPPSSSWSPGVVYIKYACFDDTISLHYCNRELRIAEWGAESEGYIRTPGYPHFYVGDECRWRLIANPEQRIRVTFLDVSLRSIGPFENECKDFVSVQDSNGDNLLYSCEQVDMPIKLTSTTNTVEVAVEARSKGAYPKRGVLLHYKSIGCVTLSAPSSGYLVYRNEDVAHYMCNVNFVFVDTRQRARLLWCYDDNRWNNTVPLCIAETVVKAVGNDSSREGIGKETKLPPHQEPNMIVDIVIPSLLIAALFIGNAFIVLIIYKYRKRSVVIVVYGTHIVCPFQVLPNVDK
Summary
Uniprot
A0A194PZ06
A0A194R2V3
A0A2A4JXB9
A0A0L7LA75
A0A1Y1MQP8
E0VL19
+ More
U4UTM2 A0A1B0GQ86 A0A1W4X0C5 A0A026X3Y5 D6WJU5 A0A158NLT6 A0A3L8DJA5 A0A151ILC4 A0A195FV16 A0A147BG24 A0A2A3EJ58 A0A182S5P6 W5JJU0 A0A0L7R2W2 A0A182W6Y0 A0A182RCQ0 A0A1Q3FVB0 A0A1Q3FVT9 A0A1Y9GKV4 K7IQT3 A0A2R5LAI0 A0A1J1IEE0 T1IKQ1 A0A232FBC3 B7P5Z9 E9J388 A0A224YKB9 A0A224YQR1 A0A131YWS1 A0A0P5AMX8 A0A0P6IX99 A0A0P5FEA4 A0A0P5C4U2 A0A0P5BN45 E2C845 A0A0C9RL71 A0A0P6E827 A0A151HZR8 A0A1B0CNI3 A0A151WZP1 A0A151J8L2 A0A0P5N5J1 A0A0N8BXQ4 A0A154PM52 E2AQT1 A0A088A1F8 A0A0P6GDV3 E9FZR5 A0A0P5EQS8 A0A0N8C3M2 A0A182F0R2 A0A0N8DR70 A0A0M9A5L7 A0A0P4YP61 A0A0P4YDL3 A0A2L2YDK1 A0A1D1VM48 A0A1Y3BG07 A0A1V9XBT4 A0A0K2UAV1 A0A0K2UBP3 A0A0K2UAV4 T1KS84 A0A0P5C4X7 A0A0P5XG60 A0A0P5KKP3
U4UTM2 A0A1B0GQ86 A0A1W4X0C5 A0A026X3Y5 D6WJU5 A0A158NLT6 A0A3L8DJA5 A0A151ILC4 A0A195FV16 A0A147BG24 A0A2A3EJ58 A0A182S5P6 W5JJU0 A0A0L7R2W2 A0A182W6Y0 A0A182RCQ0 A0A1Q3FVB0 A0A1Q3FVT9 A0A1Y9GKV4 K7IQT3 A0A2R5LAI0 A0A1J1IEE0 T1IKQ1 A0A232FBC3 B7P5Z9 E9J388 A0A224YKB9 A0A224YQR1 A0A131YWS1 A0A0P5AMX8 A0A0P6IX99 A0A0P5FEA4 A0A0P5C4U2 A0A0P5BN45 E2C845 A0A0C9RL71 A0A0P6E827 A0A151HZR8 A0A1B0CNI3 A0A151WZP1 A0A151J8L2 A0A0P5N5J1 A0A0N8BXQ4 A0A154PM52 E2AQT1 A0A088A1F8 A0A0P6GDV3 E9FZR5 A0A0P5EQS8 A0A0N8C3M2 A0A182F0R2 A0A0N8DR70 A0A0M9A5L7 A0A0P4YP61 A0A0P4YDL3 A0A2L2YDK1 A0A1D1VM48 A0A1Y3BG07 A0A1V9XBT4 A0A0K2UAV1 A0A0K2UBP3 A0A0K2UAV4 T1KS84 A0A0P5C4X7 A0A0P5XG60 A0A0P5KKP3
Pubmed
EMBL
KQ459584
KPI98556.1
KQ460855
KPJ11849.1
NWSH01000448
PCG76338.1
+ More
JTDY01002089 KOB72136.1 GEZM01030354 GEZM01030349 JAV85607.1 DS235263 EEB14075.1 KB632364 ERL93541.1 AJVK01006584 AJVK01006585 KK107019 EZA62706.1 KQ971343 EFA03096.2 ADTU01019826 ADTU01019827 ADTU01019828 ADTU01019829 ADTU01019830 ADTU01019831 QOIP01000007 RLU20019.1 KQ977121 KYN05528.1 KQ981241 KYN44271.1 GEGO01005693 JAR89711.1 KZ288250 PBC31061.1 ADMH02001286 ETN63170.1 KQ414664 KOC65168.1 GFDL01003511 JAV31534.1 GFDL01003512 JAV31533.1 APCN01002463 APCN01002464 GGLE01002408 MBY06534.1 CVRI01000047 CRK97366.1 JH430595 NNAY01000475 OXU28126.1 ABJB010152318 ABJB010720239 ABJB011013757 DS643202 EEC02021.1 GL768059 EFZ12705.1 GFPF01006952 MAA18098.1 GFPF01006953 MAA18099.1 GEDV01005931 JAP82626.1 GDIP01201485 JAJ21917.1 GDIQ01026407 JAN68330.1 GDIP01148851 JAJ74551.1 GDIP01175542 JAJ47860.1 GDIP01187295 LRGB01001581 JAJ36107.1 KZS11422.1 GL453538 EFN75898.1 GBYB01008990 GBYB01011737 JAG78757.1 JAG81504.1 GDIQ01078247 JAN16490.1 KQ976699 KYM77432.1 AJWK01020377 AJWK01020378 AJWK01020379 AJWK01020380 KQ982635 KYQ53339.1 KQ979548 KYN21085.1 GDIQ01150816 JAL00910.1 GDIQ01134627 JAL17099.1 KQ434978 KZC12961.1 GL441834 EFN64193.1 GDIQ01054411 JAN40326.1 GL732528 EFX87101.1 GDIP01143135 JAJ80267.1 GDIQ01118083 JAL33643.1 GDIP01008887 JAM94828.1 KQ435748 KOX76433.1 GDIP01224583 JAI98818.1 GDIP01232580 JAI90821.1 IAAA01018067 IAAA01018068 IAAA01018069 LAA05430.1 BDGG01000008 GAV02690.1 MUJZ01021360 OTF79792.1 MNPL01015596 OQR71005.1 HACA01017982 CDW35343.1 HACA01017981 CDW35342.1 HACA01017983 CDW35344.1 CAEY01000423 GDIP01175541 JAJ47861.1 GDIP01072761 JAM30954.1 GDIQ01182775 JAK68950.1
JTDY01002089 KOB72136.1 GEZM01030354 GEZM01030349 JAV85607.1 DS235263 EEB14075.1 KB632364 ERL93541.1 AJVK01006584 AJVK01006585 KK107019 EZA62706.1 KQ971343 EFA03096.2 ADTU01019826 ADTU01019827 ADTU01019828 ADTU01019829 ADTU01019830 ADTU01019831 QOIP01000007 RLU20019.1 KQ977121 KYN05528.1 KQ981241 KYN44271.1 GEGO01005693 JAR89711.1 KZ288250 PBC31061.1 ADMH02001286 ETN63170.1 KQ414664 KOC65168.1 GFDL01003511 JAV31534.1 GFDL01003512 JAV31533.1 APCN01002463 APCN01002464 GGLE01002408 MBY06534.1 CVRI01000047 CRK97366.1 JH430595 NNAY01000475 OXU28126.1 ABJB010152318 ABJB010720239 ABJB011013757 DS643202 EEC02021.1 GL768059 EFZ12705.1 GFPF01006952 MAA18098.1 GFPF01006953 MAA18099.1 GEDV01005931 JAP82626.1 GDIP01201485 JAJ21917.1 GDIQ01026407 JAN68330.1 GDIP01148851 JAJ74551.1 GDIP01175542 JAJ47860.1 GDIP01187295 LRGB01001581 JAJ36107.1 KZS11422.1 GL453538 EFN75898.1 GBYB01008990 GBYB01011737 JAG78757.1 JAG81504.1 GDIQ01078247 JAN16490.1 KQ976699 KYM77432.1 AJWK01020377 AJWK01020378 AJWK01020379 AJWK01020380 KQ982635 KYQ53339.1 KQ979548 KYN21085.1 GDIQ01150816 JAL00910.1 GDIQ01134627 JAL17099.1 KQ434978 KZC12961.1 GL441834 EFN64193.1 GDIQ01054411 JAN40326.1 GL732528 EFX87101.1 GDIP01143135 JAJ80267.1 GDIQ01118083 JAL33643.1 GDIP01008887 JAM94828.1 KQ435748 KOX76433.1 GDIP01224583 JAI98818.1 GDIP01232580 JAI90821.1 IAAA01018067 IAAA01018068 IAAA01018069 LAA05430.1 BDGG01000008 GAV02690.1 MUJZ01021360 OTF79792.1 MNPL01015596 OQR71005.1 HACA01017982 CDW35343.1 HACA01017981 CDW35342.1 HACA01017983 CDW35344.1 CAEY01000423 GDIP01175541 JAJ47861.1 GDIP01072761 JAM30954.1 GDIQ01182775 JAK68950.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000037510
UP000009046
UP000030742
+ More
UP000092462 UP000192223 UP000053097 UP000007266 UP000005205 UP000279307 UP000078542 UP000078541 UP000242457 UP000075901 UP000000673 UP000053825 UP000075920 UP000075900 UP000075840 UP000002358 UP000183832 UP000215335 UP000001555 UP000076858 UP000008237 UP000078540 UP000092461 UP000075809 UP000078492 UP000076502 UP000000311 UP000005203 UP000000305 UP000069272 UP000053105 UP000186922 UP000192247 UP000015104
UP000092462 UP000192223 UP000053097 UP000007266 UP000005205 UP000279307 UP000078542 UP000078541 UP000242457 UP000075901 UP000000673 UP000053825 UP000075920 UP000075900 UP000075840 UP000002358 UP000183832 UP000215335 UP000001555 UP000076858 UP000008237 UP000078540 UP000092461 UP000075809 UP000078492 UP000076502 UP000000311 UP000005203 UP000000305 UP000069272 UP000053105 UP000186922 UP000192247 UP000015104
Interpro
Gene 3D
ProteinModelPortal
A0A194PZ06
A0A194R2V3
A0A2A4JXB9
A0A0L7LA75
A0A1Y1MQP8
E0VL19
+ More
U4UTM2 A0A1B0GQ86 A0A1W4X0C5 A0A026X3Y5 D6WJU5 A0A158NLT6 A0A3L8DJA5 A0A151ILC4 A0A195FV16 A0A147BG24 A0A2A3EJ58 A0A182S5P6 W5JJU0 A0A0L7R2W2 A0A182W6Y0 A0A182RCQ0 A0A1Q3FVB0 A0A1Q3FVT9 A0A1Y9GKV4 K7IQT3 A0A2R5LAI0 A0A1J1IEE0 T1IKQ1 A0A232FBC3 B7P5Z9 E9J388 A0A224YKB9 A0A224YQR1 A0A131YWS1 A0A0P5AMX8 A0A0P6IX99 A0A0P5FEA4 A0A0P5C4U2 A0A0P5BN45 E2C845 A0A0C9RL71 A0A0P6E827 A0A151HZR8 A0A1B0CNI3 A0A151WZP1 A0A151J8L2 A0A0P5N5J1 A0A0N8BXQ4 A0A154PM52 E2AQT1 A0A088A1F8 A0A0P6GDV3 E9FZR5 A0A0P5EQS8 A0A0N8C3M2 A0A182F0R2 A0A0N8DR70 A0A0M9A5L7 A0A0P4YP61 A0A0P4YDL3 A0A2L2YDK1 A0A1D1VM48 A0A1Y3BG07 A0A1V9XBT4 A0A0K2UAV1 A0A0K2UBP3 A0A0K2UAV4 T1KS84 A0A0P5C4X7 A0A0P5XG60 A0A0P5KKP3
U4UTM2 A0A1B0GQ86 A0A1W4X0C5 A0A026X3Y5 D6WJU5 A0A158NLT6 A0A3L8DJA5 A0A151ILC4 A0A195FV16 A0A147BG24 A0A2A3EJ58 A0A182S5P6 W5JJU0 A0A0L7R2W2 A0A182W6Y0 A0A182RCQ0 A0A1Q3FVB0 A0A1Q3FVT9 A0A1Y9GKV4 K7IQT3 A0A2R5LAI0 A0A1J1IEE0 T1IKQ1 A0A232FBC3 B7P5Z9 E9J388 A0A224YKB9 A0A224YQR1 A0A131YWS1 A0A0P5AMX8 A0A0P6IX99 A0A0P5FEA4 A0A0P5C4U2 A0A0P5BN45 E2C845 A0A0C9RL71 A0A0P6E827 A0A151HZR8 A0A1B0CNI3 A0A151WZP1 A0A151J8L2 A0A0P5N5J1 A0A0N8BXQ4 A0A154PM52 E2AQT1 A0A088A1F8 A0A0P6GDV3 E9FZR5 A0A0P5EQS8 A0A0N8C3M2 A0A182F0R2 A0A0N8DR70 A0A0M9A5L7 A0A0P4YP61 A0A0P4YDL3 A0A2L2YDK1 A0A1D1VM48 A0A1Y3BG07 A0A1V9XBT4 A0A0K2UAV1 A0A0K2UBP3 A0A0K2UAV4 T1KS84 A0A0P5C4X7 A0A0P5XG60 A0A0P5KKP3
PDB
6GJE
E-value=0.00102657,
Score=99
Ontologies
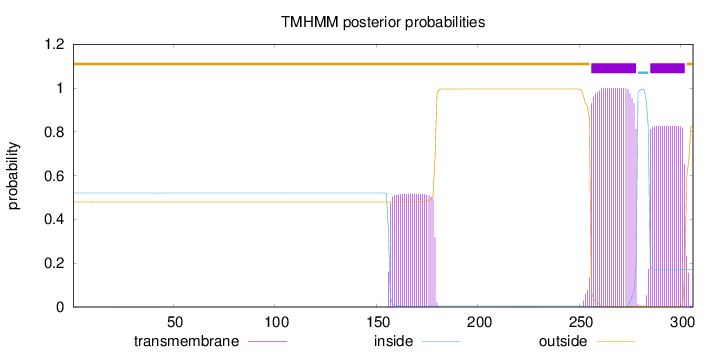
Topology
Length:
306
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
49.89512
Exp number, first 60 AAs:
0.00949
Total prob of N-in:
0.52011
outside
1 - 255
TMhelix
256 - 278
inside
279 - 284
TMhelix
285 - 302
outside
303 - 306
Population Genetic Test Statistics
Pi
270.822909
Theta
167.302099
Tajima's D
2.150404
CLR
0.414281
CSRT
0.907404629768512
Interpretation
Uncertain
