Pre Gene Modal
BGIBMGA006016
Annotation
PREDICTED:_lysosomal_thioesterase_PPT2_homolog_[Bombyx_mori]
Full name
Lysosomal thioesterase PPT2 homolog
Location in the cell
Nuclear Reliability : 2.743
Sequence
CDS
ATGAATCTTGTTAAAGCTTTATTGTTTAATGTATTAAGTTTAAGTATAGGATTTGCGTATAAGCCTGTGGTTTTAATGCATGGAATTATGACTGGGAGTGGTAGTATGGAAATAATAAAGCATAGAATAGAAGAGAAACATCCCGGAACAGTTGTGTACAATGTGAACAGGTTCGAGAGCTGGTCCAGCTTGGAAAGTATGTGGCACCAAGTTTTGGAAATTGGTTTGGACGTGGCTAACATATCTGCGTCACATCCTGAAGGAATCAATTTAATAGGATATTCTCAAGGAGGTCTGGTAGCAAGAGGGATCGTCGAGACATTCTCTAATGTATCTGTTAGTACATTCATATCACTTAGTTCACCACAAGCTGGTCAATATGGAGCTGGTTTCCTGCACTTGGTGTTTCCGGGCTTGGTCAAAGAAACTGTCTATGAGTTGTTCTACTCGAAAGTTGGTCAGCACACATCCGTTGGCAATTACTGGAACGACCCATACCATCAGCCGCTGTACGAGACCTACTGCATGTTTTTGCCTTACCTAAATAATCATAAACAGTCGAAGAAGTCTGAAGAATTCAAAGCCAACATGCTGAGACTGAAGCGGCTGGTGCTGATCGGGGGACCGGACGACGACGTCATCACACCCTGGCAGAGCAGTCAATTCGGCTACTACAACGCCAACGAAACGGTCATCGAGCTTCGCAATCAGGAAATCTACAAGGACGACAAAATAGGACTGAAGAGTCTAGACGAGTCGGGCAGGCTACACGTCCTCAGCGTTCCAGGTGTGAACCATTTCAATTGGCACATGAACGTCAGCGTAGTCGACGACTATCTGTTGCCATATTTAGATTAG
Protein
MNLVKALLFNVLSLSIGFAYKPVVLMHGIMTGSGSMEIIKHRIEEKHPGTVVYNVNRFESWSSLESMWHQVLEIGLDVANISASHPEGINLIGYSQGGLVARGIVETFSNVSVSTFISLSSPQAGQYGAGFLHLVFPGLVKETVYELFYSKVGQHTSVGNYWNDPYHQPLYETYCMFLPYLNNHKQSKKSEEFKANMLRLKRLVLIGGPDDDVITPWQSSQFGYYNANETVIELRNQEIYKDDKIGLKSLDESGRLHVLSVPGVNHFNWHMNVSVVDDYLLPYLD
Summary
Description
Probable thioesterase removing fatty acyl groups from various substrates such as S-palmitoyl-CoA. Because of structural constraints, may be unable to remove palmitate from peptides or proteins (By similarity).
Similarity
Belongs to the palmitoyl-protein thioesterase family.
Keywords
Complete proteome
Glycoprotein
Hydrolase
Lysosome
Reference proteome
Signal
Feature
chain Lysosomal thioesterase PPT2 homolog
Uniprot
H9J924
A0A291P128
A0A291P156
A0A2A4JXH6
A0A1V0M893
A0A2H1VF96
+ More
A0A2H1VHY7 A0A194PZU0 A0A212F6T0 A0A088M9J3 A0A2A3EID9 A0A1Y1MSQ9 A0A087ZMY7 V5GM39 A0A194R7J9 J3JUM0 D7ELF0 A0A026WH65 A0A1W4XE11 A0A0N0BCW2 A0A232F5I2 E9IJU4 A0A151WYL2 A0A1W7R7K7 Q17LU4 A0A1S4EYA3 A0A195F8J3 U5ES24 B0W633 A0A158ND04 A0A1Q3FPC1 A0A067RNM5 A0A0K8TH02 A0A0A9XD97 A0A0P6IZL3 A0A2P8YZ28 E2AXZ9 A0A182F259 A0A2M4BW37 A0A2M4BW29 A0A195D0E2 A0A2M4AXL1 A0A2M4BW96 W5JHN8 A0A069DYE8 A0A182PTV0 A0A2J7QU23 A0A182YFT9 A0A1L8DPT1 A0A1L8DPP2 A0A1L8DQ35 A0A182IMY8 E2BR36 A0A182VU04 F4X0L5 A0A182QW16 A0A0L7R6C1 A0A182KFK0 A0A1B6C2B5 A0A182GKK7 A0A3B0K1N0 A0A195D8L2 A0A1B6MNE8 A0A336MGD0 A0A336L6E1 A0A146M7A1 A0A1W4VM03 B4KH97 B4N7B1 A0A182HI70 A0A182TR60 A0A1B0DKY8 A0A182UPE0 Q29M27 B4G6M7 E0VFQ7 A0A195B824 A0A182XN25 A0A1B0CR03 Q7Q1P3 Q9VKH6 A0A0M5IYF9 B4LQF9 A0A224XWU3 A0A0V0GB72 B4QAD3 B3N4J1 A0A0K8W5F9 A0A1B6HTE7 B4HX37 A0A1B6IIK6 A0A1B6K4L0 A0A182MNI9 A0A1B6I5T9 A0A1B6I7R3 B3MMV1 A0A1I8PK54 B4P1F9 A0A0C9RNF9 A0A0T6B8F2
A0A2H1VHY7 A0A194PZU0 A0A212F6T0 A0A088M9J3 A0A2A3EID9 A0A1Y1MSQ9 A0A087ZMY7 V5GM39 A0A194R7J9 J3JUM0 D7ELF0 A0A026WH65 A0A1W4XE11 A0A0N0BCW2 A0A232F5I2 E9IJU4 A0A151WYL2 A0A1W7R7K7 Q17LU4 A0A1S4EYA3 A0A195F8J3 U5ES24 B0W633 A0A158ND04 A0A1Q3FPC1 A0A067RNM5 A0A0K8TH02 A0A0A9XD97 A0A0P6IZL3 A0A2P8YZ28 E2AXZ9 A0A182F259 A0A2M4BW37 A0A2M4BW29 A0A195D0E2 A0A2M4AXL1 A0A2M4BW96 W5JHN8 A0A069DYE8 A0A182PTV0 A0A2J7QU23 A0A182YFT9 A0A1L8DPT1 A0A1L8DPP2 A0A1L8DQ35 A0A182IMY8 E2BR36 A0A182VU04 F4X0L5 A0A182QW16 A0A0L7R6C1 A0A182KFK0 A0A1B6C2B5 A0A182GKK7 A0A3B0K1N0 A0A195D8L2 A0A1B6MNE8 A0A336MGD0 A0A336L6E1 A0A146M7A1 A0A1W4VM03 B4KH97 B4N7B1 A0A182HI70 A0A182TR60 A0A1B0DKY8 A0A182UPE0 Q29M27 B4G6M7 E0VFQ7 A0A195B824 A0A182XN25 A0A1B0CR03 Q7Q1P3 Q9VKH6 A0A0M5IYF9 B4LQF9 A0A224XWU3 A0A0V0GB72 B4QAD3 B3N4J1 A0A0K8W5F9 A0A1B6HTE7 B4HX37 A0A1B6IIK6 A0A1B6K4L0 A0A182MNI9 A0A1B6I5T9 A0A1B6I7R3 B3MMV1 A0A1I8PK54 B4P1F9 A0A0C9RNF9 A0A0T6B8F2
EC Number
3.1.2.-
Pubmed
19121390
28986331
28349297
26354079
22118469
26385554
+ More
28004739 22516182 23537049 18362917 19820115 24508170 30249741 28648823 21282665 17510324 21347285 24845553 25401762 26999592 29403074 20798317 20920257 23761445 26334808 25244985 21719571 26483478 26823975 17994087 15632085 20566863 12364791 18719403 10731132 12537572 12537569 22936249 17550304
28004739 22516182 23537049 18362917 19820115 24508170 30249741 28648823 21282665 17510324 21347285 24845553 25401762 26999592 29403074 20798317 20920257 23761445 26334808 25244985 21719571 26483478 26823975 17994087 15632085 20566863 12364791 18719403 10731132 12537572 12537569 22936249 17550304
EMBL
BABH01004596
MF687649
ATJ44575.1
MF706178
ATJ44605.1
NWSH01000468
+ More
PCG76203.1 KU755493 ARD71203.1 ODYU01001984 SOQ38934.1 ODYU01002656 SOQ40443.1 KQ459584 KPI98553.1 AGBW02009976 OWR49437.1 KJ579208 AIN34684.1 KZ288242 PBC31234.1 GEZM01023912 JAV88138.1 GALX01005789 JAB62677.1 KQ460855 KPJ11846.1 APGK01021327 BT126935 KB740314 KB631743 AEE61897.1 ENN80889.1 ERL85730.1 DS497846 EFA12062.1 KK107213 QOIP01000007 EZA55298.1 RLU20575.1 KQ435896 KOX69241.1 NNAY01000988 OXU25597.1 GL763883 EFZ19122.1 KQ982650 KYQ52876.1 GEHC01000528 JAV47117.1 CH477212 EAT47669.1 KQ981744 KYN36374.1 GANO01002557 JAB57314.1 DS231846 EDS36181.1 ADTU01012128 GFDL01005595 JAV29450.1 KK852514 KDR22190.1 GBRD01008672 GBRD01001042 JAG64779.1 GBHO01024914 GBHO01024912 JAG18690.1 JAG18692.1 GDUN01000656 JAN95263.1 PYGN01000277 PSN49504.1 GL443740 EFN61702.1 GGFJ01008071 MBW57212.1 GGFJ01008073 MBW57214.1 KQ977012 KYN06385.1 GGFK01012212 MBW45533.1 GGFJ01008070 MBW57211.1 ADMH02001369 ETN62823.1 GBGD01002435 JAC86454.1 NEVH01011191 PNF32079.1 GFDF01005635 JAV08449.1 GFDF01005636 JAV08448.1 GFDF01005634 JAV08450.1 GL449898 EFN81842.1 GL888498 EGI60007.1 AXCN02000619 KQ414646 KOC66388.1 GEDC01029636 JAS07662.1 JXUM01070038 KQ562585 KXJ75544.1 OUUW01000004 SPP79506.1 KQ981135 KYN09230.1 GEBQ01002507 JAT37470.1 UFQT01001120 SSX29000.1 UFQS01002029 UFQT01002029 SSX13004.1 SSX32444.1 GDHC01008281 GDHC01002978 JAQ10348.1 JAQ15651.1 CH933807 EDW13314.1 CH964182 EDW80252.1 APCN01005638 AJVK01006563 CH379060 EAL33868.1 CH479180 EDW29141.1 DS235123 EEB12213.1 KQ976558 KYM80683.1 AJWK01024273 AAAB01008980 EAA14558.2 AE014134 BT021446 AY060722 CP012523 ALC39311.1 CH940649 EDW63409.1 GFTR01003975 JAW12451.1 GECL01000872 JAP05252.1 CM000361 CM002910 EDX04688.1 KMY89748.1 CH954177 EDV58903.1 GDHF01006027 JAI46287.1 GECU01029749 JAS77957.1 CH480818 EDW52582.1 GECU01020961 JAS86745.1 GECU01027529 GECU01021627 GECU01019621 GECU01018918 GECU01016736 GECU01004621 GECU01001317 JAS80177.1 JAS86079.1 JAS88085.1 JAS88788.1 JAS90970.1 JAT03086.1 JAT06390.1 AXCM01002101 GECU01025410 JAS82296.1 GECU01024728 JAS82978.1 CH902620 EDV30976.2 CM000157 EDW88066.2 GBYB01015037 JAG84804.1 LJIG01009170 KRT83616.1
PCG76203.1 KU755493 ARD71203.1 ODYU01001984 SOQ38934.1 ODYU01002656 SOQ40443.1 KQ459584 KPI98553.1 AGBW02009976 OWR49437.1 KJ579208 AIN34684.1 KZ288242 PBC31234.1 GEZM01023912 JAV88138.1 GALX01005789 JAB62677.1 KQ460855 KPJ11846.1 APGK01021327 BT126935 KB740314 KB631743 AEE61897.1 ENN80889.1 ERL85730.1 DS497846 EFA12062.1 KK107213 QOIP01000007 EZA55298.1 RLU20575.1 KQ435896 KOX69241.1 NNAY01000988 OXU25597.1 GL763883 EFZ19122.1 KQ982650 KYQ52876.1 GEHC01000528 JAV47117.1 CH477212 EAT47669.1 KQ981744 KYN36374.1 GANO01002557 JAB57314.1 DS231846 EDS36181.1 ADTU01012128 GFDL01005595 JAV29450.1 KK852514 KDR22190.1 GBRD01008672 GBRD01001042 JAG64779.1 GBHO01024914 GBHO01024912 JAG18690.1 JAG18692.1 GDUN01000656 JAN95263.1 PYGN01000277 PSN49504.1 GL443740 EFN61702.1 GGFJ01008071 MBW57212.1 GGFJ01008073 MBW57214.1 KQ977012 KYN06385.1 GGFK01012212 MBW45533.1 GGFJ01008070 MBW57211.1 ADMH02001369 ETN62823.1 GBGD01002435 JAC86454.1 NEVH01011191 PNF32079.1 GFDF01005635 JAV08449.1 GFDF01005636 JAV08448.1 GFDF01005634 JAV08450.1 GL449898 EFN81842.1 GL888498 EGI60007.1 AXCN02000619 KQ414646 KOC66388.1 GEDC01029636 JAS07662.1 JXUM01070038 KQ562585 KXJ75544.1 OUUW01000004 SPP79506.1 KQ981135 KYN09230.1 GEBQ01002507 JAT37470.1 UFQT01001120 SSX29000.1 UFQS01002029 UFQT01002029 SSX13004.1 SSX32444.1 GDHC01008281 GDHC01002978 JAQ10348.1 JAQ15651.1 CH933807 EDW13314.1 CH964182 EDW80252.1 APCN01005638 AJVK01006563 CH379060 EAL33868.1 CH479180 EDW29141.1 DS235123 EEB12213.1 KQ976558 KYM80683.1 AJWK01024273 AAAB01008980 EAA14558.2 AE014134 BT021446 AY060722 CP012523 ALC39311.1 CH940649 EDW63409.1 GFTR01003975 JAW12451.1 GECL01000872 JAP05252.1 CM000361 CM002910 EDX04688.1 KMY89748.1 CH954177 EDV58903.1 GDHF01006027 JAI46287.1 GECU01029749 JAS77957.1 CH480818 EDW52582.1 GECU01020961 JAS86745.1 GECU01027529 GECU01021627 GECU01019621 GECU01018918 GECU01016736 GECU01004621 GECU01001317 JAS80177.1 JAS86079.1 JAS88085.1 JAS88788.1 JAS90970.1 JAT03086.1 JAT06390.1 AXCM01002101 GECU01025410 JAS82296.1 GECU01024728 JAS82978.1 CH902620 EDV30976.2 CM000157 EDW88066.2 GBYB01015037 JAG84804.1 LJIG01009170 KRT83616.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000242457
UP000005203
+ More
UP000053240 UP000019118 UP000030742 UP000007266 UP000053097 UP000279307 UP000192223 UP000053105 UP000215335 UP000075809 UP000008820 UP000078541 UP000002320 UP000005205 UP000027135 UP000245037 UP000000311 UP000069272 UP000078542 UP000000673 UP000075885 UP000235965 UP000076408 UP000075880 UP000008237 UP000075920 UP000007755 UP000075886 UP000053825 UP000075881 UP000069940 UP000249989 UP000268350 UP000078492 UP000192221 UP000009192 UP000007798 UP000075840 UP000075902 UP000092462 UP000075903 UP000001819 UP000008744 UP000009046 UP000078540 UP000076407 UP000092461 UP000007062 UP000000803 UP000092553 UP000008792 UP000000304 UP000008711 UP000001292 UP000075883 UP000007801 UP000095300 UP000002282
UP000053240 UP000019118 UP000030742 UP000007266 UP000053097 UP000279307 UP000192223 UP000053105 UP000215335 UP000075809 UP000008820 UP000078541 UP000002320 UP000005205 UP000027135 UP000245037 UP000000311 UP000069272 UP000078542 UP000000673 UP000075885 UP000235965 UP000076408 UP000075880 UP000008237 UP000075920 UP000007755 UP000075886 UP000053825 UP000075881 UP000069940 UP000249989 UP000268350 UP000078492 UP000192221 UP000009192 UP000007798 UP000075840 UP000075902 UP000092462 UP000075903 UP000001819 UP000008744 UP000009046 UP000078540 UP000076407 UP000092461 UP000007062 UP000000803 UP000092553 UP000008792 UP000000304 UP000008711 UP000001292 UP000075883 UP000007801 UP000095300 UP000002282
Pfam
PF02089 Palm_thioest
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9J924
A0A291P128
A0A291P156
A0A2A4JXH6
A0A1V0M893
A0A2H1VF96
+ More
A0A2H1VHY7 A0A194PZU0 A0A212F6T0 A0A088M9J3 A0A2A3EID9 A0A1Y1MSQ9 A0A087ZMY7 V5GM39 A0A194R7J9 J3JUM0 D7ELF0 A0A026WH65 A0A1W4XE11 A0A0N0BCW2 A0A232F5I2 E9IJU4 A0A151WYL2 A0A1W7R7K7 Q17LU4 A0A1S4EYA3 A0A195F8J3 U5ES24 B0W633 A0A158ND04 A0A1Q3FPC1 A0A067RNM5 A0A0K8TH02 A0A0A9XD97 A0A0P6IZL3 A0A2P8YZ28 E2AXZ9 A0A182F259 A0A2M4BW37 A0A2M4BW29 A0A195D0E2 A0A2M4AXL1 A0A2M4BW96 W5JHN8 A0A069DYE8 A0A182PTV0 A0A2J7QU23 A0A182YFT9 A0A1L8DPT1 A0A1L8DPP2 A0A1L8DQ35 A0A182IMY8 E2BR36 A0A182VU04 F4X0L5 A0A182QW16 A0A0L7R6C1 A0A182KFK0 A0A1B6C2B5 A0A182GKK7 A0A3B0K1N0 A0A195D8L2 A0A1B6MNE8 A0A336MGD0 A0A336L6E1 A0A146M7A1 A0A1W4VM03 B4KH97 B4N7B1 A0A182HI70 A0A182TR60 A0A1B0DKY8 A0A182UPE0 Q29M27 B4G6M7 E0VFQ7 A0A195B824 A0A182XN25 A0A1B0CR03 Q7Q1P3 Q9VKH6 A0A0M5IYF9 B4LQF9 A0A224XWU3 A0A0V0GB72 B4QAD3 B3N4J1 A0A0K8W5F9 A0A1B6HTE7 B4HX37 A0A1B6IIK6 A0A1B6K4L0 A0A182MNI9 A0A1B6I5T9 A0A1B6I7R3 B3MMV1 A0A1I8PK54 B4P1F9 A0A0C9RNF9 A0A0T6B8F2
A0A2H1VHY7 A0A194PZU0 A0A212F6T0 A0A088M9J3 A0A2A3EID9 A0A1Y1MSQ9 A0A087ZMY7 V5GM39 A0A194R7J9 J3JUM0 D7ELF0 A0A026WH65 A0A1W4XE11 A0A0N0BCW2 A0A232F5I2 E9IJU4 A0A151WYL2 A0A1W7R7K7 Q17LU4 A0A1S4EYA3 A0A195F8J3 U5ES24 B0W633 A0A158ND04 A0A1Q3FPC1 A0A067RNM5 A0A0K8TH02 A0A0A9XD97 A0A0P6IZL3 A0A2P8YZ28 E2AXZ9 A0A182F259 A0A2M4BW37 A0A2M4BW29 A0A195D0E2 A0A2M4AXL1 A0A2M4BW96 W5JHN8 A0A069DYE8 A0A182PTV0 A0A2J7QU23 A0A182YFT9 A0A1L8DPT1 A0A1L8DPP2 A0A1L8DQ35 A0A182IMY8 E2BR36 A0A182VU04 F4X0L5 A0A182QW16 A0A0L7R6C1 A0A182KFK0 A0A1B6C2B5 A0A182GKK7 A0A3B0K1N0 A0A195D8L2 A0A1B6MNE8 A0A336MGD0 A0A336L6E1 A0A146M7A1 A0A1W4VM03 B4KH97 B4N7B1 A0A182HI70 A0A182TR60 A0A1B0DKY8 A0A182UPE0 Q29M27 B4G6M7 E0VFQ7 A0A195B824 A0A182XN25 A0A1B0CR03 Q7Q1P3 Q9VKH6 A0A0M5IYF9 B4LQF9 A0A224XWU3 A0A0V0GB72 B4QAD3 B3N4J1 A0A0K8W5F9 A0A1B6HTE7 B4HX37 A0A1B6IIK6 A0A1B6K4L0 A0A182MNI9 A0A1B6I5T9 A0A1B6I7R3 B3MMV1 A0A1I8PK54 B4P1F9 A0A0C9RNF9 A0A0T6B8F2
PDB
1PJA
E-value=1.39974e-58,
Score=571
Ontologies
PATHWAY
PANTHER
Topology
Subcellular location
Lysosome
SignalP
Position: 1 - 19,
Likelihood: 0.628333
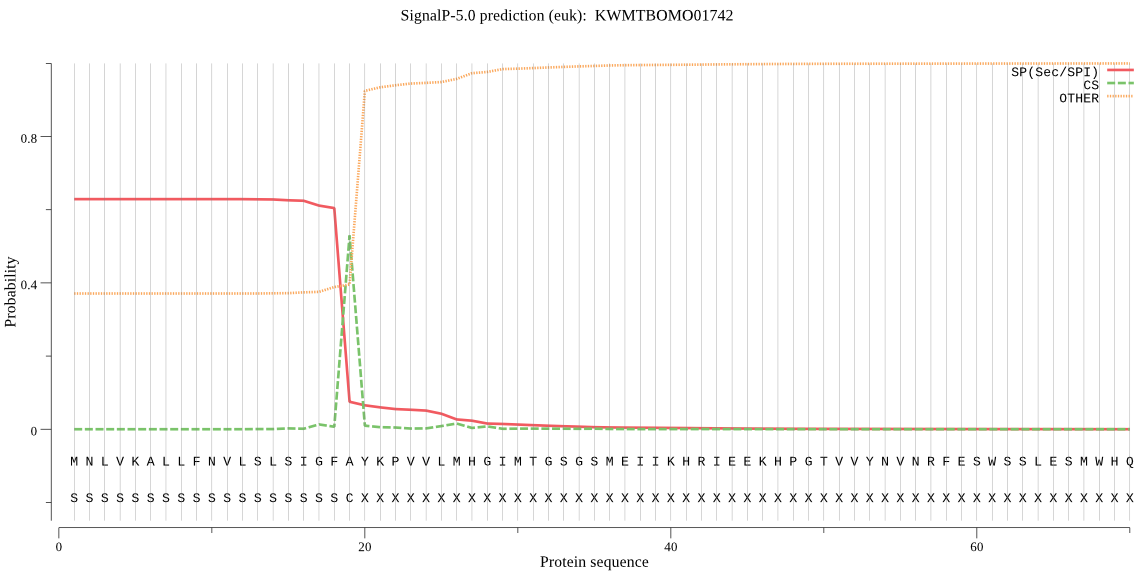
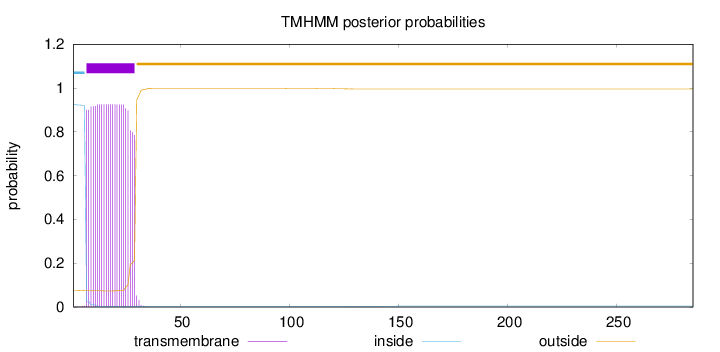
Length:
285
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.93684
Exp number, first 60 AAs:
20.88955
Total prob of N-in:
0.92511
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 285
Population Genetic Test Statistics
Pi
188.145105
Theta
170.484268
Tajima's D
-0.458964
CLR
283.703168
CSRT
0.248587570621469
Interpretation
Uncertain
