Gene
KWMTBOMO01730
Pre Gene Modal
BGIBMGA006131
Annotation
ICE_protein_[Bombyx_mori]
Full name
Caspase-3
+ More
Caspase
Caspase
Alternative Name
Apopain
Cysteine protease CPP32
drICE
Cysteine protease CPP32
drICE
Location in the cell
Nuclear Reliability : 1.2
Sequence
CDS
ATGGAGGAGACTTCGAACGATTTCAAAATTTTCAACGCGAACCCATCCAATTCGAGCAATAAAGAAGTCATGGATAACTCAGAAGAGACGCAACCGACAGTGAGTTTTGACCCGGAATCAGTGTATTACAATATGTCCGGAGATAAATACTTGATTATCTTCAATCATTACACCTTCAAGAAAACCAAATACTTTCAATATAAACAGCCCTCGACAAGGAACGGTACATTTGAAGATGTTAAAAGTTTAAAGGATTTATTCGAAGGTCTATCTTACAATGTTACAACTTACACCGATCTTATCTATGAAGATATTTTAAATAGAGTTGCTGAATTTTGCAAGCGCGATCATAGCAAGACCTCATGTGTTATTGTGACGATCCTGACACATGGCGATAGCGGCGAAGTATTCGCTGCCGACCAACCATACAAGATCAGCGACCTCACTAATATGATTGAGAATGCTCATCACACGCTAGTCGGCAAGCCTAAGATCTTTTTTATTCAGGCATGCCGTGGTCCCTTTATGGATGAGGGCAGAACAGTGGTGCTGGATGGTGAAAGCAGGGGCGTCTTACGCCTTCCAACGCATTCGGACTTCCTCTTCTTACATTCTTCTGTTGACGGTTATCTTTCCTACCGTGATTCAAGAGGATCGTGGCTAATCCAGACACTGTGCGAAATCATTAGAAAGAATCACAAGGAAATCGATCTACTGCATATGATCACGATGGTGAACAAAAGCATTGCATATGCGAAGAGTACGTACGTAAAGCAGATGACCGAAACTAGGTTTACTTTAACTAAGTTATTTATGTTTGAAAAGTAG
Protein
MEETSNDFKIFNANPSNSSNKEVMDNSEETQPTVSFDPESVYYNMSGDKYLIIFNHYTFKKTKYFQYKQPSTRNGTFEDVKSLKDLFEGLSYNVTTYTDLIYEDILNRVAEFCKRDHSKTSCVIVTILTHGDSGEVFAADQPYKISDLTNMIENAHHTLVGKPKIFFIQACRGPFMDEGRTVVLDGESRGVLRLPTHSDFLFLHSSVDGYLSYRDSRGSWLIQTLCEIIRKNHKEIDLLHMITMVNKSIAYAKSTYVKQMTETRFTLTKLFMFEK
Summary
Description
Important mediator of apoptosis. At the onset of apoptosis it proteolytically cleaves poly(ADP-ribose) polymerase (PARP) at a '216-Asp-|-Gly-217' bond (By similarity).
Catalytic Activity
Strict requirement for an Asp residue at positions P1 and P4. It has a preferred cleavage sequence of Asp-Xaa-Xaa-Asp-|- with a hydrophobic amino-acid residue at P2 and a hydrophilic amino-acid residue at P3, although Val or Ala are also accepted at this position.
Subunit
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 17 kDa (p17) and a 12 kDa (p12) subunit.
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 21 kDa (p21) and a 12 kDa (p12) subunit. Inactive pro-form can homodimerize. Dronc and Drice can form a stable complex (PubMed:10675329). Interacts with Diap2 (via BIR3 domain) to form a stable complex (PubMed:18166655).
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 21 kDa (p21) and a 12 kDa (p12) subunit. Inactive pro-form can homodimerize. Dronc and Drice can form a stable complex (PubMed:10675329). Interacts with Diap2 (via BIR3 domain) to form a stable complex (PubMed:18166655).
Miscellaneous
The subunits are derived from the precursor sequence by a probable autocatalytic mechanism and probably by other caspases.
Similarity
Belongs to the peptidase C14A family.
Keywords
Apoptosis
Cytoplasm
Hydrolase
Protease
Thiol protease
Zymogen
3D-structure
Complete proteome
Reference proteome
Feature
chain Caspase-3 subunit p17
Uniprot
Q5EG73
H9J9D9
Q2HZ05
Q2HZ04
R4I3L3
G0XQG3
+ More
G0XQF8 G0XQF9 A0A2H1WCN5 G0XQG2 A0A2A4K9J0 I7C0U4 F6K5S4 A0A194R219 G0XQG1 A0A2H1WCX0 A0A212F1N5 A0A194Q558 G0XQF7 A0A1E1WR75 G0XQF6 A0A087UZR2 A0A1Q3F2D4 B0WPG0 A0A182GKE2 A0A1W4W9I1 A0A0K0LBN4 A0A195CTV8 A0A1W4W9I8 A0A088A747 Q16GK6 A0A293M9N4 F4WFL7 W5JU83 A0A151WIC1 A0A1P7XV02 A0A087UZQ9 A0A1S4G240 F1NV61 K4PXX6 A0A1Y1MC92 Q17FE3 H6V6K4 Q8I955 V5G2T8 A0A0M9AAM0 A0A1L8EIG3 A0A151J4A4 B0WLP6 A0A0M4EQI3 G1NEU9 B6EEC1 P55866 A0A0L0BTI1 A0A151HYK2 B4NBM0 B4R093 O01382 A0A336MXM6 A0A3B0JG01 U3JHT6 A0A1I8M9Z6 G0XQE4 A0A1L8HLJ6 A0A182YTP8 A0A1I8P6R3 F8UU16 A0A1W4UCU4 A0A0Q3PW47 Q4KLU8 B4HZF1 G0XQE9 G1ELA2 J9JS12 B4KAN6 A9Q0J3
G0XQF8 G0XQF9 A0A2H1WCN5 G0XQG2 A0A2A4K9J0 I7C0U4 F6K5S4 A0A194R219 G0XQG1 A0A2H1WCX0 A0A212F1N5 A0A194Q558 G0XQF7 A0A1E1WR75 G0XQF6 A0A087UZR2 A0A1Q3F2D4 B0WPG0 A0A182GKE2 A0A1W4W9I1 A0A0K0LBN4 A0A195CTV8 A0A1W4W9I8 A0A088A747 Q16GK6 A0A293M9N4 F4WFL7 W5JU83 A0A151WIC1 A0A1P7XV02 A0A087UZQ9 A0A1S4G240 F1NV61 K4PXX6 A0A1Y1MC92 Q17FE3 H6V6K4 Q8I955 V5G2T8 A0A0M9AAM0 A0A1L8EIG3 A0A151J4A4 B0WLP6 A0A0M4EQI3 G1NEU9 B6EEC1 P55866 A0A0L0BTI1 A0A151HYK2 B4NBM0 B4R093 O01382 A0A336MXM6 A0A3B0JG01 U3JHT6 A0A1I8M9Z6 G0XQE4 A0A1L8HLJ6 A0A182YTP8 A0A1I8P6R3 F8UU16 A0A1W4UCU4 A0A0Q3PW47 Q4KLU8 B4HZF1 G0XQE9 G1ELA2 J9JS12 B4KAN6 A9Q0J3
EC Number
3.4.22.56
3.4.22.-
3.4.22.-
Pubmed
EMBL
AY885228
AAW79564.1
BABH01004564
BABH01004565
DQ360829
ABC94941.1
+ More
DQ360830 ABC94942.1 JQ768050 AFJ04534.1 HQ328967 AEK20824.1 HQ328962 AEK20819.1 HQ328963 AEK20820.1 ODYU01007753 SOQ50797.1 HQ328966 AEK20823.1 NWSH01000036 PCG80400.1 JX040312 AFO64610.1 HM234681 AEF30499.1 KQ460855 KPJ11833.1 HQ328965 AEK20822.1 ODYU01007752 SOQ50796.1 AGBW02010853 OWR47623.1 KQ459584 KPI98545.1 HQ328961 AEK20818.1 GDQN01006847 GDQN01002154 GDQN01001576 JAT84207.1 JAT88900.1 JAT89478.1 HQ328960 AEK20817.1 KK122506 KFM82851.1 GFDL01013329 JAV21716.1 DS232023 EDS32339.1 JXUM01069817 KQ562571 KXJ75579.1 KF956388 AIX10294.1 KQ977329 KYN03569.1 CH478268 EAT33368.1 GFWV01012062 GFWV01020562 MAA36791.1 GL888120 EGI66994.1 ADMH02000120 ETN67696.1 KQ983089 KYQ47566.1 KF955542 KF955543 KZ288416 AIS23518.1 AIS23519.1 PBC26031.1 KFM82848.1 AADN05000306 AB755808 BAM62940.1 GEZM01037080 GEZM01037077 JAV82210.1 CH477271 EAT45302.1 JN794535 AFA43940.1 AF548387 AAO16241.1 GALX01004136 JAB64330.1 KQ435699 KOX80590.1 GFDG01000341 JAV18458.1 KQ980159 KYN17427.1 DS231989 EDS30611.1 CP012526 ALC47264.1 FJ266474 ACI43910.1 D89784 JRES01001482 KNC22529.1 KQ976719 KYM76754.1 CH964232 EDW81184.2 CM000364 EDX14899.1 Y12261 AE014297 AY058451 UFQT01001457 SSX30638.1 OUUW01000005 SPP81055.1 AGTO01010918 HQ328948 AEK20805.1 CM004467 OCT96911.1 JF906728 AEH76885.1 LMAW01002534 KQK80332.1 BC098991 AAH98991.1 CH480819 EDW53408.1 HQ328953 AEK20810.1 JN113041 AEK12771.1 ABLF02008613 CH933806 EDW16773.1 EF190236 ABO93468.1
DQ360830 ABC94942.1 JQ768050 AFJ04534.1 HQ328967 AEK20824.1 HQ328962 AEK20819.1 HQ328963 AEK20820.1 ODYU01007753 SOQ50797.1 HQ328966 AEK20823.1 NWSH01000036 PCG80400.1 JX040312 AFO64610.1 HM234681 AEF30499.1 KQ460855 KPJ11833.1 HQ328965 AEK20822.1 ODYU01007752 SOQ50796.1 AGBW02010853 OWR47623.1 KQ459584 KPI98545.1 HQ328961 AEK20818.1 GDQN01006847 GDQN01002154 GDQN01001576 JAT84207.1 JAT88900.1 JAT89478.1 HQ328960 AEK20817.1 KK122506 KFM82851.1 GFDL01013329 JAV21716.1 DS232023 EDS32339.1 JXUM01069817 KQ562571 KXJ75579.1 KF956388 AIX10294.1 KQ977329 KYN03569.1 CH478268 EAT33368.1 GFWV01012062 GFWV01020562 MAA36791.1 GL888120 EGI66994.1 ADMH02000120 ETN67696.1 KQ983089 KYQ47566.1 KF955542 KF955543 KZ288416 AIS23518.1 AIS23519.1 PBC26031.1 KFM82848.1 AADN05000306 AB755808 BAM62940.1 GEZM01037080 GEZM01037077 JAV82210.1 CH477271 EAT45302.1 JN794535 AFA43940.1 AF548387 AAO16241.1 GALX01004136 JAB64330.1 KQ435699 KOX80590.1 GFDG01000341 JAV18458.1 KQ980159 KYN17427.1 DS231989 EDS30611.1 CP012526 ALC47264.1 FJ266474 ACI43910.1 D89784 JRES01001482 KNC22529.1 KQ976719 KYM76754.1 CH964232 EDW81184.2 CM000364 EDX14899.1 Y12261 AE014297 AY058451 UFQT01001457 SSX30638.1 OUUW01000005 SPP81055.1 AGTO01010918 HQ328948 AEK20805.1 CM004467 OCT96911.1 JF906728 AEH76885.1 LMAW01002534 KQK80332.1 BC098991 AAH98991.1 CH480819 EDW53408.1 HQ328953 AEK20810.1 JN113041 AEK12771.1 ABLF02008613 CH933806 EDW16773.1 EF190236 ABO93468.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000054359
+ More
UP000002320 UP000069940 UP000249989 UP000192223 UP000078542 UP000005203 UP000008820 UP000007755 UP000000673 UP000075809 UP000242457 UP000000539 UP000053105 UP000078492 UP000092553 UP000001645 UP000037069 UP000078540 UP000007798 UP000000304 UP000000803 UP000268350 UP000016665 UP000095301 UP000186698 UP000076420 UP000095300 UP000192221 UP000051836 UP000001292 UP000007819 UP000009192
UP000002320 UP000069940 UP000249989 UP000192223 UP000078542 UP000005203 UP000008820 UP000007755 UP000000673 UP000075809 UP000242457 UP000000539 UP000053105 UP000078492 UP000092553 UP000001645 UP000037069 UP000078540 UP000007798 UP000000304 UP000000803 UP000268350 UP000016665 UP000095301 UP000186698 UP000076420 UP000095300 UP000192221 UP000051836 UP000001292 UP000007819 UP000009192
Interpro
SUPFAM
SSF52129
SSF52129
CDD
ProteinModelPortal
Q5EG73
H9J9D9
Q2HZ05
Q2HZ04
R4I3L3
G0XQG3
+ More
G0XQF8 G0XQF9 A0A2H1WCN5 G0XQG2 A0A2A4K9J0 I7C0U4 F6K5S4 A0A194R219 G0XQG1 A0A2H1WCX0 A0A212F1N5 A0A194Q558 G0XQF7 A0A1E1WR75 G0XQF6 A0A087UZR2 A0A1Q3F2D4 B0WPG0 A0A182GKE2 A0A1W4W9I1 A0A0K0LBN4 A0A195CTV8 A0A1W4W9I8 A0A088A747 Q16GK6 A0A293M9N4 F4WFL7 W5JU83 A0A151WIC1 A0A1P7XV02 A0A087UZQ9 A0A1S4G240 F1NV61 K4PXX6 A0A1Y1MC92 Q17FE3 H6V6K4 Q8I955 V5G2T8 A0A0M9AAM0 A0A1L8EIG3 A0A151J4A4 B0WLP6 A0A0M4EQI3 G1NEU9 B6EEC1 P55866 A0A0L0BTI1 A0A151HYK2 B4NBM0 B4R093 O01382 A0A336MXM6 A0A3B0JG01 U3JHT6 A0A1I8M9Z6 G0XQE4 A0A1L8HLJ6 A0A182YTP8 A0A1I8P6R3 F8UU16 A0A1W4UCU4 A0A0Q3PW47 Q4KLU8 B4HZF1 G0XQE9 G1ELA2 J9JS12 B4KAN6 A9Q0J3
G0XQF8 G0XQF9 A0A2H1WCN5 G0XQG2 A0A2A4K9J0 I7C0U4 F6K5S4 A0A194R219 G0XQG1 A0A2H1WCX0 A0A212F1N5 A0A194Q558 G0XQF7 A0A1E1WR75 G0XQF6 A0A087UZR2 A0A1Q3F2D4 B0WPG0 A0A182GKE2 A0A1W4W9I1 A0A0K0LBN4 A0A195CTV8 A0A1W4W9I8 A0A088A747 Q16GK6 A0A293M9N4 F4WFL7 W5JU83 A0A151WIC1 A0A1P7XV02 A0A087UZQ9 A0A1S4G240 F1NV61 K4PXX6 A0A1Y1MC92 Q17FE3 H6V6K4 Q8I955 V5G2T8 A0A0M9AAM0 A0A1L8EIG3 A0A151J4A4 B0WLP6 A0A0M4EQI3 G1NEU9 B6EEC1 P55866 A0A0L0BTI1 A0A151HYK2 B4NBM0 B4R093 O01382 A0A336MXM6 A0A3B0JG01 U3JHT6 A0A1I8M9Z6 G0XQE4 A0A1L8HLJ6 A0A182YTP8 A0A1I8P6R3 F8UU16 A0A1W4UCU4 A0A0Q3PW47 Q4KLU8 B4HZF1 G0XQE9 G1ELA2 J9JS12 B4KAN6 A9Q0J3
PDB
3SIR
E-value=8.41219e-33,
Score=349
Ontologies
PATHWAY
GO
GO:0006915
GO:0004197
GO:0097200
GO:0009411
GO:0005737
GO:0007507
GO:0016485
GO:0097153
GO:0051402
GO:0097194
GO:0004190
GO:0010623
GO:2001269
GO:0010165
GO:0046672
GO:0006508
GO:0012501
GO:0035070
GO:0048515
GO:0045476
GO:1990525
GO:0016322
GO:0035103
GO:0046668
GO:0008234
GO:0009982
GO:0006396
GO:0030529
GO:0016020
GO:0005515
GO:0003779
GO:0016021
PANTHER
Topology
Subcellular location
Cytoplasm
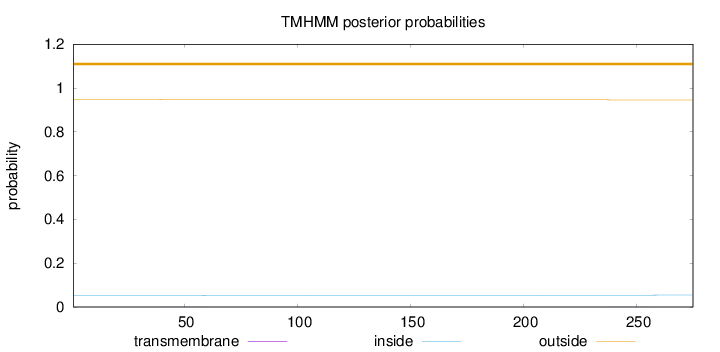
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04257
Exp number, first 60 AAs:
0.01819
Total prob of N-in:
0.05286
outside
1 - 275
Population Genetic Test Statistics
Pi
240.325572
Theta
199.263349
Tajima's D
0.870903
CLR
0.522711
CSRT
0.627668616569172
Interpretation
Uncertain
