Pre Gene Modal
BGIBMGA006020
Annotation
PREDICTED:_RNA_pseudouridylate_synthase_domain-containing_protein_2-like_isoform_X1_[Bombyx_mori]
Full name
Pseudouridine synthase
Location in the cell
Mitochondrial Reliability : 2.076
Sequence
CDS
ATGCCAATCGTGATCGTCTTAGGTAAGGGAGAAAAAGACAACGGCAAGAATATAATCTTATCTTGCGAGAAGAGGAAAGCAGATGATAGCGAAATCAACAAAGACTTAAAAAAGGCTAAATTAGAAACGAAGGCTTTAAAAGCAAAGAGGCCCGGCTTCACGGACGACAGATACAACGAGACATCATATTACATGGAAAACGGTCTTAGAAAAGTCTACCCGTACTATTTCACGTTCACGACCTTCACGAAAGGGAGATGGGTCGGGGAGAAGATCCTGGATGTCTTCGCGAGAGAGTTCAGAGCGCACCCTGCAGCGGAGTACGAAAGGTGCATCAGGGCCGGTACCCTCACGGTCAACTACGAGAGGGTTGACCCTGATTACAGACTCAAACACAACGACCTTCTAGCGAACGTAGTGCACAGGCACGAAGTACCGGTCACCAGCCAACCCATCACCCTCGTTCATATCGACGAGGACATTGTGGTTGTCAACAAACCGGCCTCTATCCCCGTGCATCCATGCGGTCGCTACAGACACAATACCGTTGTTTTTATCCTCGCGAAGGAATACAACTTGAAGAACCTGAGGACCATCCACAGACTGGACAGGCTCACGTCAGGACTTCTTCTGTTCGGGAGGAGTCCGAAGAAAGCTAGACAGATGGAGCACCAGATTAGGAACCGACAAGTCCAGAAGGAATACGTCTGCAGGGTCGACGGAGAATTTCCTGACGAGGAAATAGAATGTCAAGAGCCGATAGAAGTGGTCAGCTACAAAATAGGCGTGTGCAAGGTGTCTCCGAAAGGCAAGGACTGCTCGACTGTGTTCAAGCGACTTGGCTTCAACCAGGCGAACAACACTAGCGTGGTGCTCTGCCGCCCGAAGACCGGTAGAATGCACCAGATCAGAGTTCACCTGCAGTACCTTGGTTACCCGGTAGTAAACGATCCCCTGTACAACCACCCTGTGTTCGGTCCACTGCGCGGTAAGGGCGGCGACACCGGCGGCAAAACTGACGAGCAACTAGTCCGGGATCTGATCGCCATCCACAACGCCGAGAACTGGCTCGGTGTTGATGCTGCTGATGACGATTTGCTGTTCTCAAAGCCGGCGCCTGATGATAAAGTTGGCGAAGACGACTGCGACACAGGACCCGCTTCCAGGGAGTCTTCACCGAGATTAGATTCGCCAGCGCCCGGAGTGACAGCCTCTACCGTCATGACTCCAACTCTAGCGAGTCCGTCGAGCGGCGCTGAGGCCCCGGCGGAGGAGTCACCTCGCGCGCCACCCTCTCCCACCCCCCTCGCGACGCTCCCTATCAATGAAGACTCTAACGATGCCAAGTCAGACAAGGTGACGGTGGCGACGCAGACGGGCTGCGCGGAGACCGCAACGAGCGCGGCGGGCGCGGCTGAGGTCATCGACCCGCACTGCTACGAGTGCCGCGTGCGGTACCGCGACCCGCGCCCGCGCGACCTCGTCATGTACCTACACGCCTGGAAGTACAAGGGCCCGGGCTGGGAATACGAGACGGAGCTGCCGCAGTGGGCGAGCCTAGACTGGGACGAGCCAGAGAACTCATAG
Protein
MPIVIVLGKGEKDNGKNIILSCEKRKADDSEINKDLKKAKLETKALKAKRPGFTDDRYNETSYYMENGLRKVYPYYFTFTTFTKGRWVGEKILDVFAREFRAHPAAEYERCIRAGTLTVNYERVDPDYRLKHNDLLANVVHRHEVPVTSQPITLVHIDEDIVVVNKPASIPVHPCGRYRHNTVVFILAKEYNLKNLRTIHRLDRLTSGLLLFGRSPKKARQMEHQIRNRQVQKEYVCRVDGEFPDEEIECQEPIEVVSYKIGVCKVSPKGKDCSTVFKRLGFNQANNTSVVLCRPKTGRMHQIRVHLQYLGYPVVNDPLYNHPVFGPLRGKGGDTGGKTDEQLVRDLIAIHNAENWLGVDAADDDLLFSKPAPDDKVGEDDCDTGPASRESSPRLDSPAPGVTASTVMTPTLASPSSGAEAPAEESPRAPPSPTPLATLPINEDSNDAKSDKVTVATQTGCAETATSAAGAAEVIDPHCYECRVRYRDPRPRDLVMYLHAWKYKGPGWEYETELPQWASLDWDEPENS
Summary
Description
Responsible for synthesis of pseudouridine from uracil.
Catalytic Activity
a uridine in RNA = a psi-uridine in RNA
Similarity
Belongs to the pseudouridine synthase RluA family.
Uniprot
H9J928
A0A2A4K912
A0A2A4K7Z7
A0A182YFU0
A0A2M4CIS4
Q17LU3
+ More
Q5TP89 A0A1B6C5M5 A0A023F451 A0A1J1HFZ9 A0A146L739 D6WKG9 A0A146LH65 A0A336LNY1 A0A146M822 A0A1B0GBA4 A0A1A9UNR2 A0A1A9ZXG1 A0A336LK45 A0A1A9XTY7 A0A1B0C242 A0A1Y1LYV3 A0A146LHY8 A0A067RFR0 T1P9G1 T1HVH2 A0A1I8M359 T1PJY5 A0A3L8DHA1 B4MUA8 A0A088A1C1 B4JQ06 K7IM46 A0A026WTE9 B4LVB5 A0A182VCQ2 B4KHQ8 A0A0J9QZQ3 A0A158NNB8 A0A1L8DGS6 A0A1W4UG01 B4HWQ5 A0A2H1VEV7 B3MJJ0 B4P0P9 A0A154P6F0 B4Q9B3 A0A0R1DJT1 B3N5F0 Q9VKV0 A0A3B0JLH9 A0A0Q5WP40 Q86P70 A0A0M4E5C8 A0A194R3L9 A0A1A9W0P6 A0A1B6FQB4 A0A1B6F8K1 N6U6B2 W8BTC2 A0A034W376 A0A2S2PYJ6 A0A182NBG5 A0A0Q9W8C8 A0A0Q9W8B4 B4LVB6 A0A0A1X7N9 A0A1Q3G0D2 J9JTE7 A0A2H8TV31 A0A1B6D791 Q9VKU8 A0A0R1DR12 A0A0R1DLZ8 M9MRH0 A0A182LSB0 A0A1B6KRK1 B4HWQ6 B4MUA7 A0A1W4UST0 A0A1W4UFB8 B4G757 A0A1B6MUR0 A0A0L0CFL4 A0A0Q9W8E7 J9K377 B3N5E9 T1ISV6
Q5TP89 A0A1B6C5M5 A0A023F451 A0A1J1HFZ9 A0A146L739 D6WKG9 A0A146LH65 A0A336LNY1 A0A146M822 A0A1B0GBA4 A0A1A9UNR2 A0A1A9ZXG1 A0A336LK45 A0A1A9XTY7 A0A1B0C242 A0A1Y1LYV3 A0A146LHY8 A0A067RFR0 T1P9G1 T1HVH2 A0A1I8M359 T1PJY5 A0A3L8DHA1 B4MUA8 A0A088A1C1 B4JQ06 K7IM46 A0A026WTE9 B4LVB5 A0A182VCQ2 B4KHQ8 A0A0J9QZQ3 A0A158NNB8 A0A1L8DGS6 A0A1W4UG01 B4HWQ5 A0A2H1VEV7 B3MJJ0 B4P0P9 A0A154P6F0 B4Q9B3 A0A0R1DJT1 B3N5F0 Q9VKV0 A0A3B0JLH9 A0A0Q5WP40 Q86P70 A0A0M4E5C8 A0A194R3L9 A0A1A9W0P6 A0A1B6FQB4 A0A1B6F8K1 N6U6B2 W8BTC2 A0A034W376 A0A2S2PYJ6 A0A182NBG5 A0A0Q9W8C8 A0A0Q9W8B4 B4LVB6 A0A0A1X7N9 A0A1Q3G0D2 J9JTE7 A0A2H8TV31 A0A1B6D791 Q9VKU8 A0A0R1DR12 A0A0R1DLZ8 M9MRH0 A0A182LSB0 A0A1B6KRK1 B4HWQ6 B4MUA7 A0A1W4UST0 A0A1W4UFB8 B4G757 A0A1B6MUR0 A0A0L0CFL4 A0A0Q9W8E7 J9K377 B3N5E9 T1ISV6
EC Number
5.4.99.-
Pubmed
19121390
25244985
17510324
12364791
25474469
26823975
+ More
18362917 19820115 28004739 24845553 25315136 30249741 17994087 20075255 24508170 18057021 22936249 21347285 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26354079 23537049 24495485 25348373 25830018 26108605
18362917 19820115 28004739 24845553 25315136 30249741 17994087 20075255 24508170 18057021 22936249 21347285 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26354079 23537049 24495485 25348373 25830018 26108605
EMBL
BABH01004531
BABH01004532
BABH01004533
BABH01004534
BABH01004535
NWSH01000036
+ More
PCG80394.1 PCG80395.1 GGFL01001082 MBW65260.1 CH477212 EAT47670.1 AAAB01008980 EAL39149.3 GEDC01028495 GEDC01028378 JAS08803.1 JAS08920.1 GBBI01002456 JAC16256.1 CVRI01000001 CRK86474.1 GDHC01015647 JAQ02982.1 KQ971342 EFA03993.2 GDHC01011076 JAQ07553.1 UFQT01000037 SSX18383.1 GDHC01003723 JAQ14906.1 CCAG010011565 CCAG010011566 CCAG010011567 SSX18382.1 JXJN01024339 GEZM01043843 GEZM01043841 GEZM01043840 JAV78733.1 GDHC01011500 JAQ07129.1 KK852498 KDR22592.1 KA644548 AFP59177.1 ACPB03012938 ACPB03012939 ACPB03012940 ACPB03012941 ACPB03012942 KA649076 KA649152 AFP63705.1 QOIP01000008 RLU19696.1 CH963857 EDW76034.1 CH916372 EDV98986.1 KK107109 EZA59228.1 CH940649 EDW63294.1 KRF81019.1 CH933807 EDW13345.2 CM002910 KMY89557.1 ADTU01021209 ADTU01021210 ADTU01021211 ADTU01021212 ADTU01021213 ADTU01021214 ADTU01021215 ADTU01021216 ADTU01021217 ADTU01021218 GFDF01008519 JAV05565.1 CH480818 EDW52450.1 ODYU01002192 SOQ39365.1 CH902620 EDV32358.1 KPU73917.1 KPU73918.1 KPU73919.1 CM000157 EDW87944.1 KRJ97499.1 KQ434819 KZC06924.1 CM000361 EDX04555.1 KRJ97500.1 CH954177 EDV59029.1 KQS70601.1 AE014134 BT044578 AAF52959.2 AAF52960.2 AAN10755.1 ACI16540.1 OUUW01000006 SPP81713.1 KQS70600.1 BT003451 AAO39454.1 CP012523 ALC39284.1 KQ460855 KPJ11835.1 GECZ01023012 GECZ01017428 JAS46757.1 JAS52341.1 GECZ01023269 GECZ01012248 JAS46500.1 JAS57521.1 APGK01037835 APGK01037836 KB740949 ENN77170.1 GAMC01006327 GAMC01006325 JAC00229.1 GAKP01010759 JAC48193.1 GGMS01001237 MBY70440.1 KRF81018.1 KRF81021.1 EDW63295.2 GBXI01007542 JAD06750.1 GFDL01001777 JAV33268.1 ABLF02028880 ABLF02028881 GFXV01005995 MBW17800.1 GEDC01015768 GEDC01014515 JAS21530.1 JAS22783.1 AY058686 AAF52961.2 AAL13915.1 ADV37032.2 KRJ97504.1 KRJ97501.1 ADV37033.1 AXCM01001775 GEBQ01025926 JAT14051.1 EDW52451.1 EDW76033.1 CH479180 EDW28311.1 GEBQ01000319 JAT39658.1 JRES01000469 KNC30992.1 KRF81020.1 ABLF02015120 EDV59028.2 JH431448
PCG80394.1 PCG80395.1 GGFL01001082 MBW65260.1 CH477212 EAT47670.1 AAAB01008980 EAL39149.3 GEDC01028495 GEDC01028378 JAS08803.1 JAS08920.1 GBBI01002456 JAC16256.1 CVRI01000001 CRK86474.1 GDHC01015647 JAQ02982.1 KQ971342 EFA03993.2 GDHC01011076 JAQ07553.1 UFQT01000037 SSX18383.1 GDHC01003723 JAQ14906.1 CCAG010011565 CCAG010011566 CCAG010011567 SSX18382.1 JXJN01024339 GEZM01043843 GEZM01043841 GEZM01043840 JAV78733.1 GDHC01011500 JAQ07129.1 KK852498 KDR22592.1 KA644548 AFP59177.1 ACPB03012938 ACPB03012939 ACPB03012940 ACPB03012941 ACPB03012942 KA649076 KA649152 AFP63705.1 QOIP01000008 RLU19696.1 CH963857 EDW76034.1 CH916372 EDV98986.1 KK107109 EZA59228.1 CH940649 EDW63294.1 KRF81019.1 CH933807 EDW13345.2 CM002910 KMY89557.1 ADTU01021209 ADTU01021210 ADTU01021211 ADTU01021212 ADTU01021213 ADTU01021214 ADTU01021215 ADTU01021216 ADTU01021217 ADTU01021218 GFDF01008519 JAV05565.1 CH480818 EDW52450.1 ODYU01002192 SOQ39365.1 CH902620 EDV32358.1 KPU73917.1 KPU73918.1 KPU73919.1 CM000157 EDW87944.1 KRJ97499.1 KQ434819 KZC06924.1 CM000361 EDX04555.1 KRJ97500.1 CH954177 EDV59029.1 KQS70601.1 AE014134 BT044578 AAF52959.2 AAF52960.2 AAN10755.1 ACI16540.1 OUUW01000006 SPP81713.1 KQS70600.1 BT003451 AAO39454.1 CP012523 ALC39284.1 KQ460855 KPJ11835.1 GECZ01023012 GECZ01017428 JAS46757.1 JAS52341.1 GECZ01023269 GECZ01012248 JAS46500.1 JAS57521.1 APGK01037835 APGK01037836 KB740949 ENN77170.1 GAMC01006327 GAMC01006325 JAC00229.1 GAKP01010759 JAC48193.1 GGMS01001237 MBY70440.1 KRF81018.1 KRF81021.1 EDW63295.2 GBXI01007542 JAD06750.1 GFDL01001777 JAV33268.1 ABLF02028880 ABLF02028881 GFXV01005995 MBW17800.1 GEDC01015768 GEDC01014515 JAS21530.1 JAS22783.1 AY058686 AAF52961.2 AAL13915.1 ADV37032.2 KRJ97504.1 KRJ97501.1 ADV37033.1 AXCM01001775 GEBQ01025926 JAT14051.1 EDW52451.1 EDW76033.1 CH479180 EDW28311.1 GEBQ01000319 JAT39658.1 JRES01000469 KNC30992.1 KRF81020.1 ABLF02015120 EDV59028.2 JH431448
Proteomes
UP000005204
UP000218220
UP000076408
UP000008820
UP000007062
UP000183832
+ More
UP000007266 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000027135 UP000015103 UP000095301 UP000279307 UP000007798 UP000005203 UP000001070 UP000002358 UP000053097 UP000008792 UP000075903 UP000009192 UP000005205 UP000192221 UP000001292 UP000007801 UP000002282 UP000076502 UP000000304 UP000008711 UP000000803 UP000268350 UP000092553 UP000053240 UP000091820 UP000019118 UP000075884 UP000007819 UP000075883 UP000008744 UP000037069
UP000007266 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000027135 UP000015103 UP000095301 UP000279307 UP000007798 UP000005203 UP000001070 UP000002358 UP000053097 UP000008792 UP000075903 UP000009192 UP000005205 UP000192221 UP000001292 UP000007801 UP000002282 UP000076502 UP000000304 UP000008711 UP000000803 UP000268350 UP000092553 UP000053240 UP000091820 UP000019118 UP000075884 UP000007819 UP000075883 UP000008744 UP000037069
Pfam
PF00849 PseudoU_synth_2
Interpro
SUPFAM
SSF55120
SSF55120
CDD
ProteinModelPortal
H9J928
A0A2A4K912
A0A2A4K7Z7
A0A182YFU0
A0A2M4CIS4
Q17LU3
+ More
Q5TP89 A0A1B6C5M5 A0A023F451 A0A1J1HFZ9 A0A146L739 D6WKG9 A0A146LH65 A0A336LNY1 A0A146M822 A0A1B0GBA4 A0A1A9UNR2 A0A1A9ZXG1 A0A336LK45 A0A1A9XTY7 A0A1B0C242 A0A1Y1LYV3 A0A146LHY8 A0A067RFR0 T1P9G1 T1HVH2 A0A1I8M359 T1PJY5 A0A3L8DHA1 B4MUA8 A0A088A1C1 B4JQ06 K7IM46 A0A026WTE9 B4LVB5 A0A182VCQ2 B4KHQ8 A0A0J9QZQ3 A0A158NNB8 A0A1L8DGS6 A0A1W4UG01 B4HWQ5 A0A2H1VEV7 B3MJJ0 B4P0P9 A0A154P6F0 B4Q9B3 A0A0R1DJT1 B3N5F0 Q9VKV0 A0A3B0JLH9 A0A0Q5WP40 Q86P70 A0A0M4E5C8 A0A194R3L9 A0A1A9W0P6 A0A1B6FQB4 A0A1B6F8K1 N6U6B2 W8BTC2 A0A034W376 A0A2S2PYJ6 A0A182NBG5 A0A0Q9W8C8 A0A0Q9W8B4 B4LVB6 A0A0A1X7N9 A0A1Q3G0D2 J9JTE7 A0A2H8TV31 A0A1B6D791 Q9VKU8 A0A0R1DR12 A0A0R1DLZ8 M9MRH0 A0A182LSB0 A0A1B6KRK1 B4HWQ6 B4MUA7 A0A1W4UST0 A0A1W4UFB8 B4G757 A0A1B6MUR0 A0A0L0CFL4 A0A0Q9W8E7 J9K377 B3N5E9 T1ISV6
Q5TP89 A0A1B6C5M5 A0A023F451 A0A1J1HFZ9 A0A146L739 D6WKG9 A0A146LH65 A0A336LNY1 A0A146M822 A0A1B0GBA4 A0A1A9UNR2 A0A1A9ZXG1 A0A336LK45 A0A1A9XTY7 A0A1B0C242 A0A1Y1LYV3 A0A146LHY8 A0A067RFR0 T1P9G1 T1HVH2 A0A1I8M359 T1PJY5 A0A3L8DHA1 B4MUA8 A0A088A1C1 B4JQ06 K7IM46 A0A026WTE9 B4LVB5 A0A182VCQ2 B4KHQ8 A0A0J9QZQ3 A0A158NNB8 A0A1L8DGS6 A0A1W4UG01 B4HWQ5 A0A2H1VEV7 B3MJJ0 B4P0P9 A0A154P6F0 B4Q9B3 A0A0R1DJT1 B3N5F0 Q9VKV0 A0A3B0JLH9 A0A0Q5WP40 Q86P70 A0A0M4E5C8 A0A194R3L9 A0A1A9W0P6 A0A1B6FQB4 A0A1B6F8K1 N6U6B2 W8BTC2 A0A034W376 A0A2S2PYJ6 A0A182NBG5 A0A0Q9W8C8 A0A0Q9W8B4 B4LVB6 A0A0A1X7N9 A0A1Q3G0D2 J9JTE7 A0A2H8TV31 A0A1B6D791 Q9VKU8 A0A0R1DR12 A0A0R1DLZ8 M9MRH0 A0A182LSB0 A0A1B6KRK1 B4HWQ6 B4MUA7 A0A1W4UST0 A0A1W4UFB8 B4G757 A0A1B6MUR0 A0A0L0CFL4 A0A0Q9W8E7 J9K377 B3N5E9 T1ISV6
PDB
1XPI
E-value=3.13693e-19,
Score=235
Ontologies
KEGG
GO
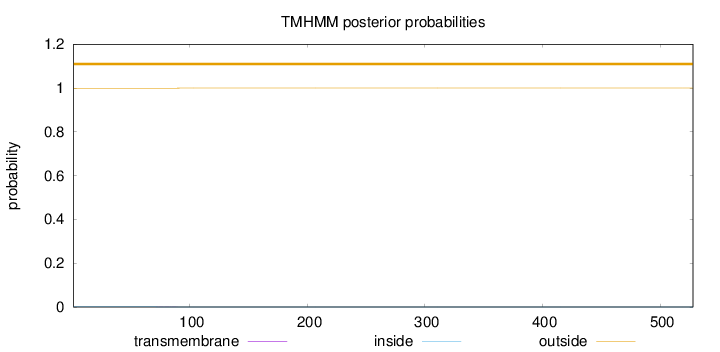
Topology
Length:
528
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0072
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00073
outside
1 - 528
Population Genetic Test Statistics
Pi
206.252109
Theta
160.845449
Tajima's D
0.940319
CLR
0.436734
CSRT
0.638818059097045
Interpretation
Uncertain
