Gene
KWMTBOMO01721
Pre Gene Modal
BGIBMGA006129
Annotation
PREDICTED:_protein_asteroid_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.552 PlasmaMembrane Reliability : 1.248
Sequence
CDS
ATGGGAGTACGAGGACTAACTACTTATATAAACAAGAATGAGGACATTTTTTTGAAAGACTTTGTATTATACGATTCCTGTGTTGTTATTGACGGTCACAGTCTCTGTGCTCAATTATATCAGCGTCTGAACAGTTTTTCTGCTTTCGGCGGCGATTATGACAAATTTTTTCAGTACGTAAGAACATTCTTTAAACATTTTCGGAAATGCAATTTAACCTCATACGTATTGTTTGACGGAAGTTACGAGACTAGAAAACTTAAAACTGCATATCATAGACTGAGATCCAAGATATATGGAGCGTCATGCCTAGATCCAGTTACTCAAGGATGCATGCAGATCTTCCCCTTGCTCATACGTAATGTTTTCAAAGAGGCATTGATTGATATTGGTGTTGCGTACACAGTATGCGAATTTGAGGCAGACGATGAAATCGCGGCTATGGCAAGGCATTTAAACTGTCCTGTACTGAGTTACGATTCAGATTTTTATATATACAACGTTTTATATATTCCATTCAACACTGTAGATTTTAAACCGAGATGTGTTATAATAGATGGTGTTAAGAAACATGTCTTAAATTGTAAAATATACAGATTTGAACAGTTAACTAATAGCTTTGAAGGAATCAAACAGGAAATTTTACCACTGCTTGCCACATTACTTGGCAATGATTTTGTGCAGAAGAGAGTTTTTGGAAAATTCTTTTCTGAATTTAAATTACCTAAGGCAAAAAAAAATCAAAATGATCAACAGAGATGTATCGAGGGCCTATTCAAATGGTTGCAGAATGAGTCTATTGATACGGCTATCTCTAAAATTATTGGAAGATTAAAGAAAAATCGCAAGGAGAAAGTATTTTATATTATTAAAAAAAGCATTGAAGGCTACAACAGAAAGAACTGTCGCTCATTGAAATACTTTAATATTCCAGAAAATACAGAAGAAATTGTTTCTGTATTACCAGAAAATGTTAATAAATTATTGGGAAAAGATTGTAATGACAATGATGACAGTGTTATACCAAATAATCAAATAGATGAAAATGAGACAATAGAAAGTCCACAATATGATTCTGATACAAATGAACAGTCGTCAGAAGAAGAGAATAGCTCTGATCAAGAAGACGATACCAATGAACTGGGTATCCCTGATTGGTTTGCAGAAGGAATAAGAAACAACACAATCCCTCACAGTAGTATAAATCTTTACACACATCATTTACATTTTGGTTCACCACAAGCTGAGGATTATAATGATGAAGATGCTTTCCTCTGTGCATTACCTATTCTGAGGTACTCTTTTGATATTCTTACAGATTACGAGCAAGAATATGTTACATATGTGAGCAGAGACAAAAGCCTCAACTATCACAGATTAATCATAGACAGAGAATATTCAATACCAAGACCTTTAGAAGTACCATATTCAAATCTTACAGATGAACAATTAAAGCTATATTTTAAAAATTTCTTCAAAATTAAAATGCCAATGTTAGATTTTAATTTAATTGATTTATTACCTAAGAGTTTCCAGTTGTATATGCTCAGCATCCTCTGGTGGGTTAACACCTGTGAAGTACCTATAGGCTTGGTTCATAGTTTATTTGTTTGCTACATTATGCTGGAGATGATTGATGAAAGCGTTGGTACAATTCGAGGTCAAAAGTATTATAATGAGAAATATCAAAAAACAATTGAAGAATACAAAAACAGAATGATTGTACTACCAAGTTTGACAGAAGGTGAATTATTTTTGAACAAAAACAAAATTCAACATGAAGACTGCTTTATAGCAGCGAAAATGCTTTTACAGTATTTCGAGATTGATGCTAAGATTAAAAAGAGACCCAAAAGCAATTATCATGTGAATAAAATACATAAATATGCCAAATTTCAGTGTTGTTTGCAACAAATTAATGATTTAAATACACTGTGTAAAAAGCCATATCAAAATACAAAGTATGGAAAGTCTTATAACGGTACCTTTGTTTATAATTTTGCACTAAAAATGGATACCCAGAATGATCCAAAAGTGTTCGTAAATCAGTATCTCAAAGATAGTACTACAATATTAATGTTTTACAATAGTCTTTGTACAGTTTATGATGAGCTGGTATTAGCGTTAGGCCTAAGACATTTTAAATGGGAGAGGTCAAAGAGCAGAAGAAAAAGAAAAGAGATGTTGGAGGATGAATTAAACTTTATTGTAAAAGGTTTTGAATCTGAAGTTGTTATATAA
Protein
MGVRGLTTYINKNEDIFLKDFVLYDSCVVIDGHSLCAQLYQRLNSFSAFGGDYDKFFQYVRTFFKHFRKCNLTSYVLFDGSYETRKLKTAYHRLRSKIYGASCLDPVTQGCMQIFPLLIRNVFKEALIDIGVAYTVCEFEADDEIAAMARHLNCPVLSYDSDFYIYNVLYIPFNTVDFKPRCVIIDGVKKHVLNCKIYRFEQLTNSFEGIKQEILPLLATLLGNDFVQKRVFGKFFSEFKLPKAKKNQNDQQRCIEGLFKWLQNESIDTAISKIIGRLKKNRKEKVFYIIKKSIEGYNRKNCRSLKYFNIPENTEEIVSVLPENVNKLLGKDCNDNDDSVIPNNQIDENETIESPQYDSDTNEQSSEEENSSDQEDDTNELGIPDWFAEGIRNNTIPHSSINLYTHHLHFGSPQAEDYNDEDAFLCALPILRYSFDILTDYEQEYVTYVSRDKSLNYHRLIIDREYSIPRPLEVPYSNLTDEQLKLYFKNFFKIKMPMLDFNLIDLLPKSFQLYMLSILWWVNTCEVPIGLVHSLFVCYIMLEMIDESVGTIRGQKYYNEKYQKTIEEYKNRMIVLPSLTEGELFLNKNKIQHEDCFIAAKMLLQYFEIDAKIKKRPKSNYHVNKIHKYAKFQCCLQQINDLNTLCKKPYQNTKYGKSYNGTFVYNFALKMDTQNDPKVFVNQYLKDSTTILMFYNSLCTVYDELVLALGLRHFKWERSKSRRKRKEMLEDELNFIVKGFESEVVI
Summary
Uniprot
H9J9D7
A0A2A4K9R4
A0A1E1WK91
A0A0L7L341
A0A194Q0M2
A0A212F1N0
+ More
A0A2J7QBV1 A0A2J7QBU7 D6WJ78 A0A1B6DNA6 A0A0L7R8M1 A0A067QXW0 E2BK26 E9J1N3 A0A151J8T7 A0A151WV60 B4MZQ1 A0A2A3E3T6 A0A158P3W3 K7IYR4 A0A232EHJ2 A0A195CNY2 A0A1W4W987 A0A1B0G8W8 A0A2M4BE05 F4WKV5 A0A154PNA7 A0A151JVJ1 A0A023EZ07 A0A195BKR7 A0A3L8DPW3 Q172X9 T1HPC6 A0A224XJ26 A0A182GF33 B4P301 Q29LI4 B3MN84 B4ID45 A0A0J9QTN0 A0A3B0K4M5 A0A182RRI9 Q7Q1V9 A0A0A1X7Y4 A0A026W1C0 A0A182JNG6 A0A1B6G3H4 A0A0A9XEL9 A0A0K8T5C4 A0A210PE65 T1JN56 E9H293 A0A146M4V6 A0A1B6DTK4 A0A146M1Q8
A0A2J7QBV1 A0A2J7QBU7 D6WJ78 A0A1B6DNA6 A0A0L7R8M1 A0A067QXW0 E2BK26 E9J1N3 A0A151J8T7 A0A151WV60 B4MZQ1 A0A2A3E3T6 A0A158P3W3 K7IYR4 A0A232EHJ2 A0A195CNY2 A0A1W4W987 A0A1B0G8W8 A0A2M4BE05 F4WKV5 A0A154PNA7 A0A151JVJ1 A0A023EZ07 A0A195BKR7 A0A3L8DPW3 Q172X9 T1HPC6 A0A224XJ26 A0A182GF33 B4P301 Q29LI4 B3MN84 B4ID45 A0A0J9QTN0 A0A3B0K4M5 A0A182RRI9 Q7Q1V9 A0A0A1X7Y4 A0A026W1C0 A0A182JNG6 A0A1B6G3H4 A0A0A9XEL9 A0A0K8T5C4 A0A210PE65 T1JN56 E9H293 A0A146M4V6 A0A1B6DTK4 A0A146M1Q8
Pubmed
EMBL
BABH01004521
NWSH01000036
PCG80402.1
GDQN01003631
JAT87423.1
JTDY01003326
+ More
KOB69736.1 KQ459584 KPI98544.1 AGBW02010853 OWR47624.1 NEVH01016296 PNF26061.1 PNF26060.1 KQ971343 EFA04435.1 GEDC01010168 JAS27130.1 KQ414632 KOC67189.1 KK853129 KDR10993.1 GL448740 EFN83934.1 GL767674 EFZ13214.1 KQ979480 KYN21258.1 KQ982708 KYQ51809.1 CH963920 EDW77836.1 KZ288405 PBC26154.1 ADTU01008606 NNAY01004486 OXU17826.1 KQ977513 KYN02187.1 CCAG010006989 GGFJ01001907 MBW51048.1 GL888206 EGI65197.1 KQ434998 KZC13329.1 KQ981701 KYN37474.1 GBBI01004277 JAC14435.1 KQ976453 KYM85320.1 QOIP01000005 RLU22401.1 CH477429 EAT41100.1 ACPB03014846 GFTR01007966 JAW08460.1 JXUM01010374 KQ560305 KXJ83147.1 CM000157 EDW87208.1 CH379060 EAL34061.1 CH902620 EDV31041.1 CH480829 EDW45471.1 CM002910 KMY87411.1 OUUW01000004 SPP79901.1 AAAB01008980 EAA14355.3 GBXI01007477 JAD06815.1 KK107570 EZA48859.1 GECZ01012780 JAS56989.1 GBHO01026346 JAG17258.1 GBRD01005087 JAG60734.1 NEDP02076750 OWF34778.1 JH431429 GL732585 EFX74129.1 GDHC01003811 JAQ14818.1 GEDC01008300 JAS28998.1 GDHC01004888 JAQ13741.1
KOB69736.1 KQ459584 KPI98544.1 AGBW02010853 OWR47624.1 NEVH01016296 PNF26061.1 PNF26060.1 KQ971343 EFA04435.1 GEDC01010168 JAS27130.1 KQ414632 KOC67189.1 KK853129 KDR10993.1 GL448740 EFN83934.1 GL767674 EFZ13214.1 KQ979480 KYN21258.1 KQ982708 KYQ51809.1 CH963920 EDW77836.1 KZ288405 PBC26154.1 ADTU01008606 NNAY01004486 OXU17826.1 KQ977513 KYN02187.1 CCAG010006989 GGFJ01001907 MBW51048.1 GL888206 EGI65197.1 KQ434998 KZC13329.1 KQ981701 KYN37474.1 GBBI01004277 JAC14435.1 KQ976453 KYM85320.1 QOIP01000005 RLU22401.1 CH477429 EAT41100.1 ACPB03014846 GFTR01007966 JAW08460.1 JXUM01010374 KQ560305 KXJ83147.1 CM000157 EDW87208.1 CH379060 EAL34061.1 CH902620 EDV31041.1 CH480829 EDW45471.1 CM002910 KMY87411.1 OUUW01000004 SPP79901.1 AAAB01008980 EAA14355.3 GBXI01007477 JAD06815.1 KK107570 EZA48859.1 GECZ01012780 JAS56989.1 GBHO01026346 JAG17258.1 GBRD01005087 JAG60734.1 NEDP02076750 OWF34778.1 JH431429 GL732585 EFX74129.1 GDHC01003811 JAQ14818.1 GEDC01008300 JAS28998.1 GDHC01004888 JAQ13741.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000007151
UP000235965
+ More
UP000007266 UP000053825 UP000027135 UP000008237 UP000078492 UP000075809 UP000007798 UP000242457 UP000005205 UP000002358 UP000215335 UP000078542 UP000192221 UP000092444 UP000007755 UP000076502 UP000078541 UP000078540 UP000279307 UP000008820 UP000015103 UP000069940 UP000249989 UP000002282 UP000001819 UP000007801 UP000001292 UP000268350 UP000075900 UP000007062 UP000053097 UP000075881 UP000242188 UP000000305
UP000007266 UP000053825 UP000027135 UP000008237 UP000078492 UP000075809 UP000007798 UP000242457 UP000005205 UP000002358 UP000215335 UP000078542 UP000192221 UP000092444 UP000007755 UP000076502 UP000078541 UP000078540 UP000279307 UP000008820 UP000015103 UP000069940 UP000249989 UP000002282 UP000001819 UP000007801 UP000001292 UP000268350 UP000075900 UP000007062 UP000053097 UP000075881 UP000242188 UP000000305
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J9D7
A0A2A4K9R4
A0A1E1WK91
A0A0L7L341
A0A194Q0M2
A0A212F1N0
+ More
A0A2J7QBV1 A0A2J7QBU7 D6WJ78 A0A1B6DNA6 A0A0L7R8M1 A0A067QXW0 E2BK26 E9J1N3 A0A151J8T7 A0A151WV60 B4MZQ1 A0A2A3E3T6 A0A158P3W3 K7IYR4 A0A232EHJ2 A0A195CNY2 A0A1W4W987 A0A1B0G8W8 A0A2M4BE05 F4WKV5 A0A154PNA7 A0A151JVJ1 A0A023EZ07 A0A195BKR7 A0A3L8DPW3 Q172X9 T1HPC6 A0A224XJ26 A0A182GF33 B4P301 Q29LI4 B3MN84 B4ID45 A0A0J9QTN0 A0A3B0K4M5 A0A182RRI9 Q7Q1V9 A0A0A1X7Y4 A0A026W1C0 A0A182JNG6 A0A1B6G3H4 A0A0A9XEL9 A0A0K8T5C4 A0A210PE65 T1JN56 E9H293 A0A146M4V6 A0A1B6DTK4 A0A146M1Q8
A0A2J7QBV1 A0A2J7QBU7 D6WJ78 A0A1B6DNA6 A0A0L7R8M1 A0A067QXW0 E2BK26 E9J1N3 A0A151J8T7 A0A151WV60 B4MZQ1 A0A2A3E3T6 A0A158P3W3 K7IYR4 A0A232EHJ2 A0A195CNY2 A0A1W4W987 A0A1B0G8W8 A0A2M4BE05 F4WKV5 A0A154PNA7 A0A151JVJ1 A0A023EZ07 A0A195BKR7 A0A3L8DPW3 Q172X9 T1HPC6 A0A224XJ26 A0A182GF33 B4P301 Q29LI4 B3MN84 B4ID45 A0A0J9QTN0 A0A3B0K4M5 A0A182RRI9 Q7Q1V9 A0A0A1X7Y4 A0A026W1C0 A0A182JNG6 A0A1B6G3H4 A0A0A9XEL9 A0A0K8T5C4 A0A210PE65 T1JN56 E9H293 A0A146M4V6 A0A1B6DTK4 A0A146M1Q8
Ontologies
GO
PANTHER
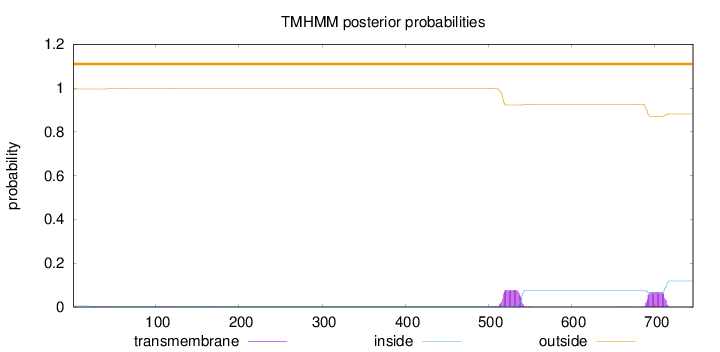
Topology
Length:
746
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.38416
Exp number, first 60 AAs:
0.03924
Total prob of N-in:
0.00478
outside
1 - 746
Population Genetic Test Statistics
Pi
37.417838
Theta
61.324625
Tajima's D
-1.181424
CLR
303.788177
CSRT
0.106694665266737
Interpretation
Uncertain
