Pre Gene Modal
BGIBMGA006025
Annotation
PREDICTED:_maestro_heat-like_repeat-containing_protein_family_member_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.291
Sequence
CDS
ATGAACACGGAGTGTCCGCAGCTGCGGGAACATTCCATTCGTCTGTTCGGCGTGCTGGCGGCGCGCGTGCGGTCCGACGCGCTGGTCGACCAGGCCGTGAACTCGCTGCCGTGCTTCTTGCTGCACCTGTGCGACAACAACCCCGCCGTGGTCCGGGCGAGCAAATTCACCTTGAAACAAGTGTTCAAGATCTTCAACGTGAAGAAATCGAACGACCTGGTGCAGACGCACCTGCTGGACGAGGGCCGCCTGTACCTGGACGAGTTCCTGTCGGCCCTGCTGCGGCAGCTGGCCGACGAGCTGCCCTCCAGCGTCGCCAGCTGTCTGCGCGCCGCCGCCCACTACCTGCACTGCGCGCGCGACGAGCTCAGGCCGCACGCGCCCACGCTGCTGGGTCTGCTCTACGCGGAACTGCTCCGCATCAGAGACAAGTACCCCGAGGACACGGAGCTGGAGCTGGACGTGTCGCGGTCGGCTCGCGCGCGCCTCCTGCAGCTGATCAAGGACAGCAGCCCCGCGGTCAGGCAAAGCTCCGCCATGGCGCTGGCGAACCTGTGCCTCGTCATCGCTTCGGACGAAATGAACGCGCAATAA
Protein
MNTECPQLREHSIRLFGVLAARVRSDALVDQAVNSLPCFLLHLCDNNPAVVRASKFTLKQVFKIFNVKKSNDLVQTHLLDEGRLYLDEFLSALLRQLADELPSSVASCLRAAAHYLHCARDELRPHAPTLLGLLYAELLRIRDKYPEDTELELDVSRSARARLLQLIKDSSPAVRQSSAMALANLCLVIASDEMNAQ
Summary
Uniprot
H9J933
A0A1E1WDY3
A0A2H1V7K3
A0A2A4K813
A0A194R6S1
A0A194PZS5
+ More
A0A212F1L6 A0A2P8XTX3 A0A1Y1NDE4 A0A146LV36 A0A0A9XNY2 A0A0A9XWL7 A0A0K8STS1 A0A2J7QBP8 A0A2S2P2B8 A0A2H8TM24 T1HVH1 A0A023EXV5 A0A182GCA6 A0A1B6MSC3 A0A1B6KA60 B0WQW2 J9LU48 A0A1B6DAP1 A0A1S4F6S6 A0A182TZ90 Q17DT8 A0A1B6HGY9 A0A2M3Z241 A0A369SKK0 A0A0V0G660 B3RJ38 A0A1D5PK39 A0A1Q3FY26 A0A0A1XR61 A0A182XBY9 A0A0A1XKM4 A0A0A1XQ34 A0A1B6FYS9 A0A182QHL8 A0A182KBM8 A0A182PSV0 A0A182L876 A0NCH7 D6WKM3 A0A182IAJ7 A0A182VGJ8 A0A182N8G4 A0A2S2QW36 A0A182W5M5 A0A182RHL1 A0A182YH71 A0A034V2Q2 A0A0K8WJ06 A0A182IWY3 A0A2P6KU33 W8B438 A0A1J1JAL4 A0A2M4B8M9 A0A2T7NJG9 A0A182FIS8 A0A2R5LMZ7 A0A2S2N4M0 A0A182M7B2 A0A0L8G9H0 T1JN14 A0A1L8FU70 A0A3Q0HHQ5 A0A3Q0HEB3 W5JHA7 A0A3Q0J9H4 H3B4H9 A0A1I8N7K6 A0A084VNW0 T1P876 A0A1I8QDF9 A0A2C9KZ46 N6SYN8 U4UQS7 A0A226MFM9 A0A218UAN6 A0A1W4YG88 A0A0L8GAE9 A0A0P5DUG9 A0A0P5NK42 A0A0N8D0H8
A0A212F1L6 A0A2P8XTX3 A0A1Y1NDE4 A0A146LV36 A0A0A9XNY2 A0A0A9XWL7 A0A0K8STS1 A0A2J7QBP8 A0A2S2P2B8 A0A2H8TM24 T1HVH1 A0A023EXV5 A0A182GCA6 A0A1B6MSC3 A0A1B6KA60 B0WQW2 J9LU48 A0A1B6DAP1 A0A1S4F6S6 A0A182TZ90 Q17DT8 A0A1B6HGY9 A0A2M3Z241 A0A369SKK0 A0A0V0G660 B3RJ38 A0A1D5PK39 A0A1Q3FY26 A0A0A1XR61 A0A182XBY9 A0A0A1XKM4 A0A0A1XQ34 A0A1B6FYS9 A0A182QHL8 A0A182KBM8 A0A182PSV0 A0A182L876 A0NCH7 D6WKM3 A0A182IAJ7 A0A182VGJ8 A0A182N8G4 A0A2S2QW36 A0A182W5M5 A0A182RHL1 A0A182YH71 A0A034V2Q2 A0A0K8WJ06 A0A182IWY3 A0A2P6KU33 W8B438 A0A1J1JAL4 A0A2M4B8M9 A0A2T7NJG9 A0A182FIS8 A0A2R5LMZ7 A0A2S2N4M0 A0A182M7B2 A0A0L8G9H0 T1JN14 A0A1L8FU70 A0A3Q0HHQ5 A0A3Q0HEB3 W5JHA7 A0A3Q0J9H4 H3B4H9 A0A1I8N7K6 A0A084VNW0 T1P876 A0A1I8QDF9 A0A2C9KZ46 N6SYN8 U4UQS7 A0A226MFM9 A0A218UAN6 A0A1W4YG88 A0A0L8GAE9 A0A0P5DUG9 A0A0P5NK42 A0A0N8D0H8
Pubmed
EMBL
BABH01004510
BABH01004511
BABH01004512
GDQN01005907
JAT85147.1
ODYU01001082
+ More
SOQ36818.1 NWSH01000036 PCG80415.1 KQ460644 KPJ13219.1 KQ459584 KPI98538.1 AGBW02010853 OWR47629.1 PYGN01001356 PSN35449.1 GEZM01009859 JAV94256.1 GDHC01008047 JAQ10582.1 GBHO01021970 JAG21634.1 GBHO01021969 JAG21635.1 GBRD01009119 JAG56702.1 NEVH01016296 PNF26007.1 GGMR01010739 MBY23358.1 GFXV01003274 MBW15079.1 ACPB03012943 GBBI01004871 JAC13841.1 JXUM01054000 JXUM01054001 KQ561800 KXJ77494.1 GEBQ01001161 JAT38816.1 GEBQ01031656 GEBQ01003499 JAT08321.1 JAT36478.1 DS232046 EDS33042.1 ABLF02034062 ABLF02034065 ABLF02034070 ABLF02034072 ABLF02042357 ABLF02046209 GEDC01020360 GEDC01014572 GEDC01013465 JAS16938.1 JAS22726.1 JAS23833.1 CH477291 EAT44574.1 GECU01033817 JAS73889.1 GGFM01001824 MBW22575.1 NOWV01000004 RDD47071.1 GECL01002912 JAP03212.1 DS985241 EDV29054.1 GFDL01002576 JAV32469.1 GBXI01001224 JAD13068.1 GBXI01002862 JAD11430.1 GBXI01000858 JAD13434.1 GECZ01014437 GECZ01001851 JAS55332.1 JAS67918.1 AXCN02002226 AAAB01008846 EAU77300.2 KQ971342 EFA03008.2 APCN01001259 GGMS01012670 MBY81873.1 GAKP01022530 JAC36422.1 GDHF01001270 JAI51044.1 MWRG01005216 PRD29835.1 GAMC01018549 GAMC01018548 JAB88006.1 CVRI01000075 CRL08614.1 GGFJ01000255 MBW49396.1 PZQS01000012 PVD21296.1 GGLE01006774 MBY10900.1 GGMT01000418 MBY12104.1 AXCM01002712 KQ423131 KOF73524.1 JH431841 CM004477 OCT75115.1 ADMH02001200 ETN63752.1 AFYH01076855 AFYH01076856 AFYH01076857 AFYH01076858 AFYH01076859 AFYH01076860 AFYH01076861 AFYH01076862 AFYH01076863 AFYH01076864 ATLV01014931 KE524996 KFB39654.1 KA644784 AFP59413.1 APGK01051854 KB741207 ENN72894.1 KB632327 ERL92511.1 MCFN01000989 OXB54094.1 MUZQ01000507 OWK50745.1 KOF73525.1 GDIP01152564 JAJ70838.1 GDIQ01151350 JAL00376.1 GDIP01080823 JAM22892.1
SOQ36818.1 NWSH01000036 PCG80415.1 KQ460644 KPJ13219.1 KQ459584 KPI98538.1 AGBW02010853 OWR47629.1 PYGN01001356 PSN35449.1 GEZM01009859 JAV94256.1 GDHC01008047 JAQ10582.1 GBHO01021970 JAG21634.1 GBHO01021969 JAG21635.1 GBRD01009119 JAG56702.1 NEVH01016296 PNF26007.1 GGMR01010739 MBY23358.1 GFXV01003274 MBW15079.1 ACPB03012943 GBBI01004871 JAC13841.1 JXUM01054000 JXUM01054001 KQ561800 KXJ77494.1 GEBQ01001161 JAT38816.1 GEBQ01031656 GEBQ01003499 JAT08321.1 JAT36478.1 DS232046 EDS33042.1 ABLF02034062 ABLF02034065 ABLF02034070 ABLF02034072 ABLF02042357 ABLF02046209 GEDC01020360 GEDC01014572 GEDC01013465 JAS16938.1 JAS22726.1 JAS23833.1 CH477291 EAT44574.1 GECU01033817 JAS73889.1 GGFM01001824 MBW22575.1 NOWV01000004 RDD47071.1 GECL01002912 JAP03212.1 DS985241 EDV29054.1 GFDL01002576 JAV32469.1 GBXI01001224 JAD13068.1 GBXI01002862 JAD11430.1 GBXI01000858 JAD13434.1 GECZ01014437 GECZ01001851 JAS55332.1 JAS67918.1 AXCN02002226 AAAB01008846 EAU77300.2 KQ971342 EFA03008.2 APCN01001259 GGMS01012670 MBY81873.1 GAKP01022530 JAC36422.1 GDHF01001270 JAI51044.1 MWRG01005216 PRD29835.1 GAMC01018549 GAMC01018548 JAB88006.1 CVRI01000075 CRL08614.1 GGFJ01000255 MBW49396.1 PZQS01000012 PVD21296.1 GGLE01006774 MBY10900.1 GGMT01000418 MBY12104.1 AXCM01002712 KQ423131 KOF73524.1 JH431841 CM004477 OCT75115.1 ADMH02001200 ETN63752.1 AFYH01076855 AFYH01076856 AFYH01076857 AFYH01076858 AFYH01076859 AFYH01076860 AFYH01076861 AFYH01076862 AFYH01076863 AFYH01076864 ATLV01014931 KE524996 KFB39654.1 KA644784 AFP59413.1 APGK01051854 KB741207 ENN72894.1 KB632327 ERL92511.1 MCFN01000989 OXB54094.1 MUZQ01000507 OWK50745.1 KOF73525.1 GDIP01152564 JAJ70838.1 GDIQ01151350 JAL00376.1 GDIP01080823 JAM22892.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000245037
+ More
UP000235965 UP000015103 UP000069940 UP000249989 UP000002320 UP000007819 UP000075902 UP000008820 UP000253843 UP000009022 UP000076407 UP000075886 UP000075881 UP000075885 UP000075882 UP000007062 UP000007266 UP000075840 UP000075903 UP000075884 UP000075920 UP000075900 UP000076408 UP000075880 UP000183832 UP000245119 UP000069272 UP000075883 UP000053454 UP000186698 UP000189705 UP000000673 UP000079169 UP000008672 UP000095301 UP000030765 UP000095300 UP000076420 UP000019118 UP000030742 UP000198323 UP000197619 UP000192224
UP000235965 UP000015103 UP000069940 UP000249989 UP000002320 UP000007819 UP000075902 UP000008820 UP000253843 UP000009022 UP000076407 UP000075886 UP000075881 UP000075885 UP000075882 UP000007062 UP000007266 UP000075840 UP000075903 UP000075884 UP000075920 UP000075900 UP000076408 UP000075880 UP000183832 UP000245119 UP000069272 UP000075883 UP000053454 UP000186698 UP000189705 UP000000673 UP000079169 UP000008672 UP000095301 UP000030765 UP000095300 UP000076420 UP000019118 UP000030742 UP000198323 UP000197619 UP000192224
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J933
A0A1E1WDY3
A0A2H1V7K3
A0A2A4K813
A0A194R6S1
A0A194PZS5
+ More
A0A212F1L6 A0A2P8XTX3 A0A1Y1NDE4 A0A146LV36 A0A0A9XNY2 A0A0A9XWL7 A0A0K8STS1 A0A2J7QBP8 A0A2S2P2B8 A0A2H8TM24 T1HVH1 A0A023EXV5 A0A182GCA6 A0A1B6MSC3 A0A1B6KA60 B0WQW2 J9LU48 A0A1B6DAP1 A0A1S4F6S6 A0A182TZ90 Q17DT8 A0A1B6HGY9 A0A2M3Z241 A0A369SKK0 A0A0V0G660 B3RJ38 A0A1D5PK39 A0A1Q3FY26 A0A0A1XR61 A0A182XBY9 A0A0A1XKM4 A0A0A1XQ34 A0A1B6FYS9 A0A182QHL8 A0A182KBM8 A0A182PSV0 A0A182L876 A0NCH7 D6WKM3 A0A182IAJ7 A0A182VGJ8 A0A182N8G4 A0A2S2QW36 A0A182W5M5 A0A182RHL1 A0A182YH71 A0A034V2Q2 A0A0K8WJ06 A0A182IWY3 A0A2P6KU33 W8B438 A0A1J1JAL4 A0A2M4B8M9 A0A2T7NJG9 A0A182FIS8 A0A2R5LMZ7 A0A2S2N4M0 A0A182M7B2 A0A0L8G9H0 T1JN14 A0A1L8FU70 A0A3Q0HHQ5 A0A3Q0HEB3 W5JHA7 A0A3Q0J9H4 H3B4H9 A0A1I8N7K6 A0A084VNW0 T1P876 A0A1I8QDF9 A0A2C9KZ46 N6SYN8 U4UQS7 A0A226MFM9 A0A218UAN6 A0A1W4YG88 A0A0L8GAE9 A0A0P5DUG9 A0A0P5NK42 A0A0N8D0H8
A0A212F1L6 A0A2P8XTX3 A0A1Y1NDE4 A0A146LV36 A0A0A9XNY2 A0A0A9XWL7 A0A0K8STS1 A0A2J7QBP8 A0A2S2P2B8 A0A2H8TM24 T1HVH1 A0A023EXV5 A0A182GCA6 A0A1B6MSC3 A0A1B6KA60 B0WQW2 J9LU48 A0A1B6DAP1 A0A1S4F6S6 A0A182TZ90 Q17DT8 A0A1B6HGY9 A0A2M3Z241 A0A369SKK0 A0A0V0G660 B3RJ38 A0A1D5PK39 A0A1Q3FY26 A0A0A1XR61 A0A182XBY9 A0A0A1XKM4 A0A0A1XQ34 A0A1B6FYS9 A0A182QHL8 A0A182KBM8 A0A182PSV0 A0A182L876 A0NCH7 D6WKM3 A0A182IAJ7 A0A182VGJ8 A0A182N8G4 A0A2S2QW36 A0A182W5M5 A0A182RHL1 A0A182YH71 A0A034V2Q2 A0A0K8WJ06 A0A182IWY3 A0A2P6KU33 W8B438 A0A1J1JAL4 A0A2M4B8M9 A0A2T7NJG9 A0A182FIS8 A0A2R5LMZ7 A0A2S2N4M0 A0A182M7B2 A0A0L8G9H0 T1JN14 A0A1L8FU70 A0A3Q0HHQ5 A0A3Q0HEB3 W5JHA7 A0A3Q0J9H4 H3B4H9 A0A1I8N7K6 A0A084VNW0 T1P876 A0A1I8QDF9 A0A2C9KZ46 N6SYN8 U4UQS7 A0A226MFM9 A0A218UAN6 A0A1W4YG88 A0A0L8GAE9 A0A0P5DUG9 A0A0P5NK42 A0A0N8D0H8
Ontologies
KEGG
GO
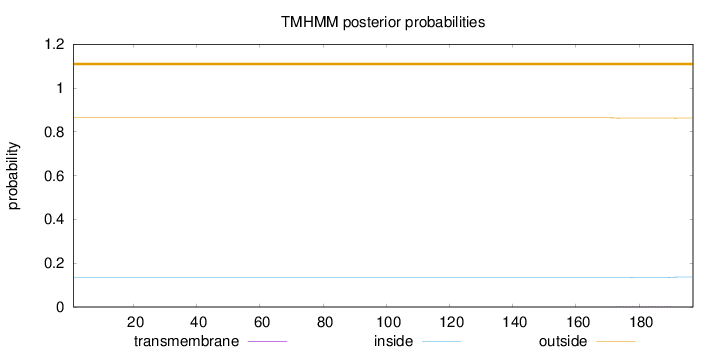
Topology
Length:
197
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03619
Exp number, first 60 AAs:
0.0023
Total prob of N-in:
0.13593
outside
1 - 197
Population Genetic Test Statistics
Pi
143.354687
Theta
140.128992
Tajima's D
0.156423
CLR
341.885354
CSRT
0.413929303534823
Interpretation
Uncertain
