Gene
KWMTBOMO01709
Pre Gene Modal
BGIBMGA006123
Annotation
PREDICTED:_endonuclease/exonuclease/phosphatase_family_domain-containing_protein_1-like_[Papilio_machaon]
Full name
Endonuclease/exonuclease/phosphatase family domain-containing protein 1
Location in the cell
Mitochondrial Reliability : 2.292 Nuclear Reliability : 2.005
Sequence
CDS
ATGGGGCAAAGTCCGAGCTCAATGAGGAACAAGAACGGTCGACACCGATCGTTCAGGAGTTTCGTAAGGCGAAGCAAATTGAACAAATCTAGCTTGAGCCATACGTTCAGTTTGCCGCCTTCAGAGGAGTATCCGGAGCTAATGAACATCAATGCAGCAACTGAGGAACAGCTGATGACCCTTCCGGGCGTGAACAGGCAACTGGCTCGAGAGATAGTACGCCACAGACAGATGATAGGTCGTTTTAAGAGAGTTGATGACCTTGCATTAGTCTCAGGTATTGGAGCTGATAAACTAGAATTACTGAGACCAGAAATATGTACGCACACAAGGAAACAGTTGTCACGGGCCAGCTCCTGTGCACACTCATTAGATAGCATACGTTTGCCCTCTGAAAGCAGACTATGTTCTATCAATTCATCCAGTGTGTTCCAACTGCAGTGTGTGCCTGGACTCAACCAAGAAATAGCTGCAAATATTGTAGATTACCGTAATAGAAAAGGGCCTTTCAAATCATTGGATGATTTAATAAAAGTTCGTGGTATGGATATTGTCAGGCTAAGTACAGTAAAACAACACTTGAGCCTGGAGTTACGTAAATCTGAAAGTGTCCAGCACTTAACTAATGGCCATGTAAATGGATGGAATGATGATAACATGCTCCAACAAAATGGAATTTCTCCAAAGACGCCACATCGAAAAAGTTTATCAATGCCAACAAAATTGACGTTACCCAATGGCTTTGCCACTGCTCCAGTAAATGATATTCTAGATTTACTCTCAGCTTATTCACATAGGCCAGTTGTAGAGGAAGTTTTCACATATGAAAGAGATGGTATTCGGTGCTGTCGACTGGCATCTTGGAATCTTCATCAGCTTAGTATAGACAAAATATCGAATCCTGGAGTGAGAGAGGTTATTTGTCGAACCATTTTAGAAAACAAATTATCGCTAATAGCCATCCAAGACATATTGAACGAGGCTGCTCTTAAAGTAATATGCGATGAGCTGAATTCTCCCGTACTAAGAAGGGTCAAAGAGTGGAAGAACAATGGGCACAAGTGGGAGTATCGTGTTTCAGGGACAAAGAACAGCCAACAACTCGGTTTTGTCTACGACTCGTCGCTGAAGGGGGTCACGGTTGAAGATATAGAGCTGGACCAGGTCTGTTTGCTGACCGAGGCCAGCGACATCAGAAGGCTTCTCTCCGAGCTGACCGCGTTGAGGAGCGACAGATATACTGCTGAGCCGCAGGCATTCCTGCTCTGCGGCAGGCCGCTGATAGCCGTGAACGTGATGTGCAGAGATCGGCTCAGCGCCGCAGACTGCGACCGGCTGGCGACCGTGGCGGCTCTAGCGGAATCCACAAAACTGAACGTGGTATTCATGGGCGAATTCCGAAACAAAGAAAACGTTCTCCAATTGAAATCGTATCAATCGATCCTTGACGAAGACTTGCCGACGACCGTGGACGCGAACGCGCTCGGCCAAAGTTTTATTTTATGTCCTGGCAACATTGAAACGAGCGGCTTCAACGGCCACTCGGGAGCCGTCAAAACTGGGTTGTGCCATCTGGCAATACCGCGAGGCTGGTCGTGGGGCGGGCCCGCGTCGCCCTTCTGTCCCATCTGGGCCGAACTCAAAGTTCCAGATTGA
Protein
MGQSPSSMRNKNGRHRSFRSFVRRSKLNKSSLSHTFSLPPSEEYPELMNINAATEEQLMTLPGVNRQLAREIVRHRQMIGRFKRVDDLALVSGIGADKLELLRPEICTHTRKQLSRASSCAHSLDSIRLPSESRLCSINSSSVFQLQCVPGLNQEIAANIVDYRNRKGPFKSLDDLIKVRGMDIVRLSTVKQHLSLELRKSESVQHLTNGHVNGWNDDNMLQQNGISPKTPHRKSLSMPTKLTLPNGFATAPVNDILDLLSAYSHRPVVEEVFTYERDGIRCCRLASWNLHQLSIDKISNPGVREVICRTILENKLSLIAIQDILNEAALKVICDELNSPVLRRVKEWKNNGHKWEYRVSGTKNSQQLGFVYDSSLKGVTVEDIELDQVCLLTEASDIRRLLSELTALRSDRYTAEPQAFLLCGRPLIAVNVMCRDRLSAADCDRLATVAALAESTKLNVVFMGEFRNKENVLQLKSYQSILDEDLPTTVDANALGQSFILCPGNIETSGFNGHSGAVKTGLCHLAIPRGWSWGGPASPFCPIWAELKVPD
Summary
Keywords
Complete proteome
Lipoprotein
Myristate
Phosphoprotein
Polymorphism
Reference proteome
Feature
chain Endonuclease/exonuclease/phosphatase family domain-containing protein 1
sequence variant In dbSNP:rs196586.
sequence variant In dbSNP:rs196586.
Uniprot
A0A2A4K902
A0A2A4K839
A0A212F1N1
A0A194R6C6
A0A194Q0L2
D6WM33
+ More
A0A1Y1KPQ0 A0A067R0L0 A0A2J7QBP3 U4UFI9 N6ULH6 A0A1B0C9M2 A0A1L8DQ98 A0A1L8DQC5 A0A1B6CQC6 A0A2P8XG88 A0A2J7QBQ2 A0A1B6JDK5 A0A087U0E5 A0A0V0GBW2 U5EZN7 A0A023EWW0 T1ISV2 A0A2P6LCC4 A0A0P4WH03 A0A0A9WQ49 A0A0A9WGQ5 A0A1D2NCU1 A0A0P4VY66 A0A2T7NGZ2 A0A0C9RHX9 A0A226F3J0 H3BHH1 A0A0P6J4J6 G5BAX9 W5P3Z5 A0A2J8UNP6 A0A2I3MLA8 G1QN22 G1SNA6 F7IHT9 A0A2K5HLP8 Q7L9B9 A0A091D7X8 A0A2K5MK69 A0A2K6BQD6 A0A2K5VK53 F7G8W8 F7CZ53 A0A2K5YVP1 G3QR97 A0A2R9BXX1 A0A0D9RR75 A0A2J8JDY3 Q3TGW2 G7MLH6 A0A2U3ZYA5 A0A2K6S0K3 A0A2K5QLF2 L8IXJ7 Q3MHJ7 A0A1D5NVM3 Q5XI74 A0A287BQI4 A0A2K5EMG9 A0A2K6KWL4 H0UVM7 A0A0Q3RA47 G3HTJ3 A0A384A4W9 G3TL67 I3MBP6 W5MJF4 A0A2Y9F995 A0A093HMA1 A0A099Z4L1 M3W817 A0A2K6QJC6 A0A087QQU9 R0LXR4 U3IQ82 H0XIT8 A0A210R0T8 A0A2J8UNQ1 A0A2K6GIP3 A0A1V4J5U1 A0A1U7QGM0 D2HA03 A0A2I0LZG9 A0A2Y9JME7 A0A1S3GQP6 A0A091GJF9 G1L427 A0A151P2I8
A0A1Y1KPQ0 A0A067R0L0 A0A2J7QBP3 U4UFI9 N6ULH6 A0A1B0C9M2 A0A1L8DQ98 A0A1L8DQC5 A0A1B6CQC6 A0A2P8XG88 A0A2J7QBQ2 A0A1B6JDK5 A0A087U0E5 A0A0V0GBW2 U5EZN7 A0A023EWW0 T1ISV2 A0A2P6LCC4 A0A0P4WH03 A0A0A9WQ49 A0A0A9WGQ5 A0A1D2NCU1 A0A0P4VY66 A0A2T7NGZ2 A0A0C9RHX9 A0A226F3J0 H3BHH1 A0A0P6J4J6 G5BAX9 W5P3Z5 A0A2J8UNP6 A0A2I3MLA8 G1QN22 G1SNA6 F7IHT9 A0A2K5HLP8 Q7L9B9 A0A091D7X8 A0A2K5MK69 A0A2K6BQD6 A0A2K5VK53 F7G8W8 F7CZ53 A0A2K5YVP1 G3QR97 A0A2R9BXX1 A0A0D9RR75 A0A2J8JDY3 Q3TGW2 G7MLH6 A0A2U3ZYA5 A0A2K6S0K3 A0A2K5QLF2 L8IXJ7 Q3MHJ7 A0A1D5NVM3 Q5XI74 A0A287BQI4 A0A2K5EMG9 A0A2K6KWL4 H0UVM7 A0A0Q3RA47 G3HTJ3 A0A384A4W9 G3TL67 I3MBP6 W5MJF4 A0A2Y9F995 A0A093HMA1 A0A099Z4L1 M3W817 A0A2K6QJC6 A0A087QQU9 R0LXR4 U3IQ82 H0XIT8 A0A210R0T8 A0A2J8UNQ1 A0A2K6GIP3 A0A1V4J5U1 A0A1U7QGM0 D2HA03 A0A2I0LZG9 A0A2Y9JME7 A0A1S3GQP6 A0A091GJF9 G1L427 A0A151P2I8
Pubmed
22118469
26354079
18362917
19820115
28004739
24845553
+ More
23537049 29403074 25474469 25401762 26823975 27289101 27129103 9215903 21993625 20809919 21993624 25243066 11214970 14702039 12690205 15489334 20213681 23186163 24275569 17431167 19892987 22398555 22722832 16136131 15368895 16141072 17242355 19144319 21183079 22002653 22751099 15592404 22673903 21804562 17975172 25362486 23749191 28812685 20010809 23371554 22293439
23537049 29403074 25474469 25401762 26823975 27289101 27129103 9215903 21993625 20809919 21993624 25243066 11214970 14702039 12690205 15489334 20213681 23186163 24275569 17431167 19892987 22398555 22722832 16136131 15368895 16141072 17242355 19144319 21183079 22002653 22751099 15592404 22673903 21804562 17975172 25362486 23749191 28812685 20010809 23371554 22293439
EMBL
NWSH01000036
PCG80398.1
PCG80397.1
AGBW02010853
OWR47631.1
KQ460644
+ More
KPJ13222.1 KQ459584 KPI98534.1 KQ971343 EFA04215.1 GEZM01078963 JAV62558.1 KK853129 KDR10992.1 NEVH01016296 PNF26010.1 KB632303 ERL91792.1 APGK01018868 APGK01018869 APGK01018870 KB740085 ENN81526.1 AJWK01002473 AJWK01002474 GFDF01005441 JAV08643.1 GFDF01005445 JAV08639.1 GEDC01021579 JAS15719.1 PYGN01002242 PSN31026.1 PNF26011.1 GECU01016822 GECU01010451 JAS90884.1 JAS97255.1 KK117576 KFM70834.1 GECL01000540 JAP05584.1 GANO01000449 JAB59422.1 GBBI01005105 JAC13607.1 AFFK01018203 JH431448 MWRG01000381 PRD36229.1 GDRN01045475 JAI66853.1 GBHO01036621 GBHO01036619 GBRD01003933 GBRD01003931 GDHC01003353 JAG06983.1 JAG06985.1 JAG61888.1 JAQ15276.1 GBHO01038076 GBHO01036620 GBRD01003934 GBRD01003932 JAG05528.1 JAG06984.1 JAG61887.1 LJIJ01000088 ODN03059.1 GDKW01002072 JAI54523.1 PZQS01000012 PVD20441.1 GBYB01007880 JAG77647.1 LNIX01000001 OXA64017.1 AFYH01010739 AFYH01010740 AFYH01010741 AFYH01010742 GEBF01004476 JAN99156.1 JH169321 EHB06440.1 AMGL01087607 AMGL01087608 AMGL01087609 AMGL01087610 ABGA01296550 ABGA01296551 ABGA01296552 ABGA01296553 ABGA01296554 ABGA01296555 ABGA01296556 NDHI03003451 PNJ46887.1 PNJ46888.1 AHZZ02022766 AHZZ02022767 AHZZ02022768 AHZZ02022769 ADFV01046492 ADFV01046493 ADFV01046494 ADFV01046495 ADFV01046496 ADFV01046497 AAGW02035771 AAGW02035772 GAMT01009820 GAMS01005256 GAMR01009683 GAMQ01002873 GAMP01007128 JAB02041.1 JAB17880.1 JAB24249.1 JAB38978.1 JAB45627.1 AB051493 AK027386 AC007327 AC078841 CH236951 CH471073 BC065518 KN123190 KFO26370.1 AQIA01046564 AQIA01046565 AQIA01046566 JSUE03028441 JSUE03028442 JSUE03028443 JSUE03028444 CABD030050753 CABD030050754 CABD030050755 CABD030050756 CABD030050757 CABD030050758 AJFE02049346 AQIB01018252 AQIB01018253 AQIB01018254 AACZ04038768 NBAG03000465 PNI20978.1 PNI20979.1 AK173239 AK009180 AK168564 BC025571 CM001255 EHH17380.1 JH880479 ELR61166.1 BC105212 AY568574 AADN05000034 BC083816 BC099214 AEMK02000111 AAKN02032248 LMAW01001969 KQK82225.1 JH000700 EGW10577.1 AGTP01012418 AGTP01012419 AGTP01012420 AGTP01012421 AGTP01012422 AGTP01012423 AGTP01012424 AGTP01012425 AHAT01018105 KL206564 KFV83778.1 KL888316 KGL75750.1 AANG04000096 KL225815 KFM03603.1 KB742702 EOB05278.1 ADON01036283 ADON01036284 AAQR03057790 AAQR03057791 AAQR03057792 AAQR03057793 AAQR03057794 AAQR03057795 AAQR03057796 NEDP02000981 OWF54572.1 PNJ46890.1 LSYS01009165 OPJ67365.1 GL192616 EFB15052.1 AKCR02000057 PKK22813.1 KL448200 KFO81284.1 ACTA01090870 ACTA01098870 ACTA01106870 AKHW03001210 KYO43223.1
KPJ13222.1 KQ459584 KPI98534.1 KQ971343 EFA04215.1 GEZM01078963 JAV62558.1 KK853129 KDR10992.1 NEVH01016296 PNF26010.1 KB632303 ERL91792.1 APGK01018868 APGK01018869 APGK01018870 KB740085 ENN81526.1 AJWK01002473 AJWK01002474 GFDF01005441 JAV08643.1 GFDF01005445 JAV08639.1 GEDC01021579 JAS15719.1 PYGN01002242 PSN31026.1 PNF26011.1 GECU01016822 GECU01010451 JAS90884.1 JAS97255.1 KK117576 KFM70834.1 GECL01000540 JAP05584.1 GANO01000449 JAB59422.1 GBBI01005105 JAC13607.1 AFFK01018203 JH431448 MWRG01000381 PRD36229.1 GDRN01045475 JAI66853.1 GBHO01036621 GBHO01036619 GBRD01003933 GBRD01003931 GDHC01003353 JAG06983.1 JAG06985.1 JAG61888.1 JAQ15276.1 GBHO01038076 GBHO01036620 GBRD01003934 GBRD01003932 JAG05528.1 JAG06984.1 JAG61887.1 LJIJ01000088 ODN03059.1 GDKW01002072 JAI54523.1 PZQS01000012 PVD20441.1 GBYB01007880 JAG77647.1 LNIX01000001 OXA64017.1 AFYH01010739 AFYH01010740 AFYH01010741 AFYH01010742 GEBF01004476 JAN99156.1 JH169321 EHB06440.1 AMGL01087607 AMGL01087608 AMGL01087609 AMGL01087610 ABGA01296550 ABGA01296551 ABGA01296552 ABGA01296553 ABGA01296554 ABGA01296555 ABGA01296556 NDHI03003451 PNJ46887.1 PNJ46888.1 AHZZ02022766 AHZZ02022767 AHZZ02022768 AHZZ02022769 ADFV01046492 ADFV01046493 ADFV01046494 ADFV01046495 ADFV01046496 ADFV01046497 AAGW02035771 AAGW02035772 GAMT01009820 GAMS01005256 GAMR01009683 GAMQ01002873 GAMP01007128 JAB02041.1 JAB17880.1 JAB24249.1 JAB38978.1 JAB45627.1 AB051493 AK027386 AC007327 AC078841 CH236951 CH471073 BC065518 KN123190 KFO26370.1 AQIA01046564 AQIA01046565 AQIA01046566 JSUE03028441 JSUE03028442 JSUE03028443 JSUE03028444 CABD030050753 CABD030050754 CABD030050755 CABD030050756 CABD030050757 CABD030050758 AJFE02049346 AQIB01018252 AQIB01018253 AQIB01018254 AACZ04038768 NBAG03000465 PNI20978.1 PNI20979.1 AK173239 AK009180 AK168564 BC025571 CM001255 EHH17380.1 JH880479 ELR61166.1 BC105212 AY568574 AADN05000034 BC083816 BC099214 AEMK02000111 AAKN02032248 LMAW01001969 KQK82225.1 JH000700 EGW10577.1 AGTP01012418 AGTP01012419 AGTP01012420 AGTP01012421 AGTP01012422 AGTP01012423 AGTP01012424 AGTP01012425 AHAT01018105 KL206564 KFV83778.1 KL888316 KGL75750.1 AANG04000096 KL225815 KFM03603.1 KB742702 EOB05278.1 ADON01036283 ADON01036284 AAQR03057790 AAQR03057791 AAQR03057792 AAQR03057793 AAQR03057794 AAQR03057795 AAQR03057796 NEDP02000981 OWF54572.1 PNJ46890.1 LSYS01009165 OPJ67365.1 GL192616 EFB15052.1 AKCR02000057 PKK22813.1 KL448200 KFO81284.1 ACTA01090870 ACTA01098870 ACTA01106870 AKHW03001210 KYO43223.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
UP000027135
+ More
UP000235965 UP000030742 UP000019118 UP000092461 UP000245037 UP000054359 UP000094527 UP000245119 UP000198287 UP000008672 UP000006813 UP000002356 UP000001595 UP000028761 UP000001073 UP000001811 UP000008225 UP000233080 UP000005640 UP000028990 UP000233060 UP000233120 UP000233100 UP000006718 UP000002281 UP000233140 UP000001519 UP000240080 UP000029965 UP000002277 UP000000589 UP000245340 UP000233220 UP000233040 UP000009136 UP000000539 UP000002494 UP000008227 UP000233020 UP000233180 UP000005447 UP000051836 UP000001075 UP000261681 UP000007646 UP000005215 UP000018468 UP000053584 UP000053641 UP000011712 UP000233200 UP000053286 UP000016666 UP000005225 UP000242188 UP000233160 UP000190648 UP000189706 UP000053872 UP000248482 UP000081671 UP000053760 UP000008912 UP000050525
UP000235965 UP000030742 UP000019118 UP000092461 UP000245037 UP000054359 UP000094527 UP000245119 UP000198287 UP000008672 UP000006813 UP000002356 UP000001595 UP000028761 UP000001073 UP000001811 UP000008225 UP000233080 UP000005640 UP000028990 UP000233060 UP000233120 UP000233100 UP000006718 UP000002281 UP000233140 UP000001519 UP000240080 UP000029965 UP000002277 UP000000589 UP000245340 UP000233220 UP000233040 UP000009136 UP000000539 UP000002494 UP000008227 UP000233020 UP000233180 UP000005447 UP000051836 UP000001075 UP000261681 UP000007646 UP000005215 UP000018468 UP000053584 UP000053641 UP000011712 UP000233200 UP000053286 UP000016666 UP000005225 UP000242188 UP000233160 UP000190648 UP000189706 UP000053872 UP000248482 UP000081671 UP000053760 UP000008912 UP000050525
Pfam
PF03372 Exo_endo_phos
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K902
A0A2A4K839
A0A212F1N1
A0A194R6C6
A0A194Q0L2
D6WM33
+ More
A0A1Y1KPQ0 A0A067R0L0 A0A2J7QBP3 U4UFI9 N6ULH6 A0A1B0C9M2 A0A1L8DQ98 A0A1L8DQC5 A0A1B6CQC6 A0A2P8XG88 A0A2J7QBQ2 A0A1B6JDK5 A0A087U0E5 A0A0V0GBW2 U5EZN7 A0A023EWW0 T1ISV2 A0A2P6LCC4 A0A0P4WH03 A0A0A9WQ49 A0A0A9WGQ5 A0A1D2NCU1 A0A0P4VY66 A0A2T7NGZ2 A0A0C9RHX9 A0A226F3J0 H3BHH1 A0A0P6J4J6 G5BAX9 W5P3Z5 A0A2J8UNP6 A0A2I3MLA8 G1QN22 G1SNA6 F7IHT9 A0A2K5HLP8 Q7L9B9 A0A091D7X8 A0A2K5MK69 A0A2K6BQD6 A0A2K5VK53 F7G8W8 F7CZ53 A0A2K5YVP1 G3QR97 A0A2R9BXX1 A0A0D9RR75 A0A2J8JDY3 Q3TGW2 G7MLH6 A0A2U3ZYA5 A0A2K6S0K3 A0A2K5QLF2 L8IXJ7 Q3MHJ7 A0A1D5NVM3 Q5XI74 A0A287BQI4 A0A2K5EMG9 A0A2K6KWL4 H0UVM7 A0A0Q3RA47 G3HTJ3 A0A384A4W9 G3TL67 I3MBP6 W5MJF4 A0A2Y9F995 A0A093HMA1 A0A099Z4L1 M3W817 A0A2K6QJC6 A0A087QQU9 R0LXR4 U3IQ82 H0XIT8 A0A210R0T8 A0A2J8UNQ1 A0A2K6GIP3 A0A1V4J5U1 A0A1U7QGM0 D2HA03 A0A2I0LZG9 A0A2Y9JME7 A0A1S3GQP6 A0A091GJF9 G1L427 A0A151P2I8
A0A1Y1KPQ0 A0A067R0L0 A0A2J7QBP3 U4UFI9 N6ULH6 A0A1B0C9M2 A0A1L8DQ98 A0A1L8DQC5 A0A1B6CQC6 A0A2P8XG88 A0A2J7QBQ2 A0A1B6JDK5 A0A087U0E5 A0A0V0GBW2 U5EZN7 A0A023EWW0 T1ISV2 A0A2P6LCC4 A0A0P4WH03 A0A0A9WQ49 A0A0A9WGQ5 A0A1D2NCU1 A0A0P4VY66 A0A2T7NGZ2 A0A0C9RHX9 A0A226F3J0 H3BHH1 A0A0P6J4J6 G5BAX9 W5P3Z5 A0A2J8UNP6 A0A2I3MLA8 G1QN22 G1SNA6 F7IHT9 A0A2K5HLP8 Q7L9B9 A0A091D7X8 A0A2K5MK69 A0A2K6BQD6 A0A2K5VK53 F7G8W8 F7CZ53 A0A2K5YVP1 G3QR97 A0A2R9BXX1 A0A0D9RR75 A0A2J8JDY3 Q3TGW2 G7MLH6 A0A2U3ZYA5 A0A2K6S0K3 A0A2K5QLF2 L8IXJ7 Q3MHJ7 A0A1D5NVM3 Q5XI74 A0A287BQI4 A0A2K5EMG9 A0A2K6KWL4 H0UVM7 A0A0Q3RA47 G3HTJ3 A0A384A4W9 G3TL67 I3MBP6 W5MJF4 A0A2Y9F995 A0A093HMA1 A0A099Z4L1 M3W817 A0A2K6QJC6 A0A087QQU9 R0LXR4 U3IQ82 H0XIT8 A0A210R0T8 A0A2J8UNQ1 A0A2K6GIP3 A0A1V4J5U1 A0A1U7QGM0 D2HA03 A0A2I0LZG9 A0A2Y9JME7 A0A1S3GQP6 A0A091GJF9 G1L427 A0A151P2I8
PDB
2DUY
E-value=5.04517e-05,
Score=113
Ontologies
Topology
Subcellular location
Nucleus
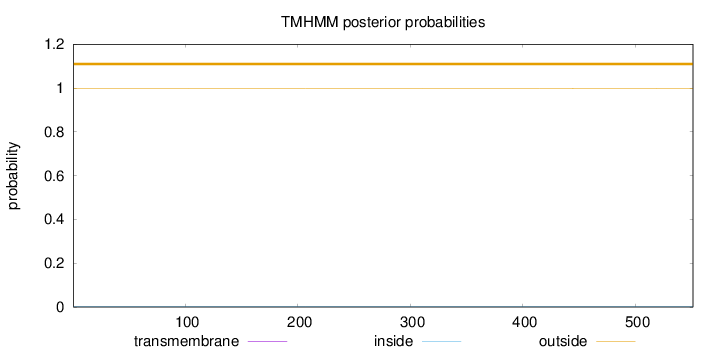
Length:
551
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00208
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00127
outside
1 - 551
Population Genetic Test Statistics
Pi
215.267969
Theta
169.214167
Tajima's D
0.759617
CLR
0.399797
CSRT
0.588620568971551
Interpretation
Uncertain
