Pre Gene Modal
BGIBMGA006121
Annotation
PREDICTED:_cytoplasmic_dynein_1_light_intermediate_chain_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.952
Sequence
CDS
ATGGATACAAATGGCCAGGCTAACGGGCTTAAATCAAAAAAGAAAGAGAACGGAGACGGCAAAGATAATTTATGGTCTGCTATATTAGAAGAAGTCCAGAATCAAGGGAACACAAAACTACCGTCAAATAAAAATGTCTTAGTACTGGGTGACAACGAAACTGGAAAAACTACCCTAGTGGCAAAGTTGCAAGGTGTTGAAGACCCAAAGAAGGGGTCTGCACTCGAATATGCGTATATAGATGTTAGAGATGAATATCGCGATGATCACACTCGGCTCAGTGTTTGGGTGCTGGATGGTGATCCAGGTCACACTAATCTACTCAAGTTTGCGCTCAGTGAGGAAACATTCCCACACACCCTAGTTATACTCACTGTGGCCATGACCACACCATGGGGTATACTAGACCAGCTCCAAAGCTGGGCTTCTGTACTTGGAGACCATATAGACAAATTAAATTTAACACCTGAACACAGGTTACAAAGTAAAAAGCAACAAGTCCAAAAATGGCAGAGGTATACGGAGCCAGGAGACGAAATGGAAGCGTCAGCTTCTTCGCCTATGAAAAGATCATCTAGAAACTTGTCGGATGATCTTGAAAATGGCGATGATGACGACAGCCCCCTGCCGGAAGCAGTACTTACTACTAATTTAGGTCTAGATATAGTGGTCGTTGTTACCAAGACTGACTACATGAGCACTCTGGAGAAGGAGCACGATTACCGCGACGAGCACCTTGACTTCATGCAGCAGTGGATCCGCCGGTTCTGTCTGCAGTACGGAGCCGCCTTGTTCTACACCAGCGCCAAGGAGGACAAGAACTGTGATCTGCTGTATAAGTATCTGACGCACAGGATATACGGTCTACCGTTCAGGACGCCGGCGCTAATTGTAGAGAAAGACGCGGTTTTGATACCAGCCGGCTGGGACAGTATGAAGAAGATTAGCATACTATATGAGAACATGCAGTCATGTAAACCTGACGATTACTATAGGGATATTATTGTTCAGCCCGCTACACGCAAAAGCGGGGGCAGTCGCGAGGCGGAGGTTCAGGCGGAGGACGAGCAGACGTTCCTGCAGCGGCAGGTGGCGGCGCTGCACGCGCTGGGCGCCGCCCCCCCGCGCGCAGACTCCCCGCTGCGCGCCTCGCCCCGCCCGCACCAGTCAGAGATCGCGAGGATCGGCGTGAGTCCGGGCACGCCGGGCAACGAGGGCGTGCTGGCGAACTTCTTCAACTCGCTGCTGTACAAGAAGACGGGCGGCGGGGGCGGCGGCGCGCGGGTGGACGGCGCGGGGGGGGCGGCCGCCGCGCGCTCCGACGCCGCCGCCGAGCTCGACCGCCTCACGCGCACCAAGCGGAGCCCGCTCGACCTCAACTCGTCCGACTGCTAG
Protein
MDTNGQANGLKSKKKENGDGKDNLWSAILEEVQNQGNTKLPSNKNVLVLGDNETGKTTLVAKLQGVEDPKKGSALEYAYIDVRDEYRDDHTRLSVWVLDGDPGHTNLLKFALSEETFPHTLVILTVAMTTPWGILDQLQSWASVLGDHIDKLNLTPEHRLQSKKQQVQKWQRYTEPGDEMEASASSPMKRSSRNLSDDLENGDDDDSPLPEAVLTTNLGLDIVVVVTKTDYMSTLEKEHDYRDEHLDFMQQWIRRFCLQYGAALFYTSAKEDKNCDLLYKYLTHRIYGLPFRTPALIVEKDAVLIPAGWDSMKKISILYENMQSCKPDDYYRDIIVQPATRKSGGSREAEVQAEDEQTFLQRQVAALHALGAAPPRADSPLRASPRPHQSEIARIGVSPGTPGNEGVLANFFNSLLYKKTGGGGGGARVDGAGGAAAARSDAAAELDRLTRTKRSPLDLNSSDC
Summary
Uniprot
A0A194R5X6
A0A194PYX3
A0A212ELH9
A0A195BHI9
A0A195F4H0
A0A158NJZ5
+ More
F4WIJ5 A0A026WF57 A0A195DZB1 E9JCD1 A0A0J7L5G6 E2C6N6 A0A151XE67 E2A100 A0A1W4W5J5 A0A2J7RBA6 D6WMX7 A0A212F1M3 A0A154PPU2 A0A3L8DP12 A0A067QP10 A0A2A3E9N2 A0A088A8B6 A0A0N0BGS3 A0A1B0C851 A0A1Y1KNP7 A0A1L8E5A5 A0A0C9R7Z7 A0A336M2P4 A0A336KG51 A0A182K743 K7IRI4 A0A182UPG6 Q7QEC0 A0A182I9R1 A0A0L0C853 W8BVG6 V5GYU1 A0A182R3G4 A0A182MIH5 A0A034VTK6 A0A0A1XRA1 U5ET37 A0A182Y7J8 A0A182PPC7 A0A182Q473 T1DE60 U4U4A8 Q16RB5 A0A232EYU9 A0A0K8U9J5 A0A182W4B3 J3JTC6 A0A182MZP3 A0A1J1HP29 A0A1Q3F4A2 A0A1Q3F452 A0A1Q3F460 A0A1Q3EU60 A0A084VVR5 A0A1B6CNW6 W8B7V1 A0A1I8QAG0 A0A182J0Q9 A0A182UEL6 A0A023EUB2 T1PDU8 A0A182KY12 A0A182X575 B3MVY2 B4JJL4 B0WSX6 A0A0C9R4M3 Q9VZ20 A0A182H422 A0A3B0JWX1 A0A0R3P537 B4GXK5 B3NV95 A0A1W4V195 B4PYE4 A0A0K8TNC2 B4M2N2 A0A1B6HG70 A0A1B6F978 A0A224XG43 A0A1B6FUE1 A0A1A9WVD1 A0A0P4VTP7 B4L4H5 Q29HJ5 B4N2K3 A0A1B6ISY7 A0A069DUL0 A0A182S7P0 A0A0V0G5K0 A0A1W4UNU4 A0A1A9YP15 A0A1B0AXR0 A0A0Q9WKF4
F4WIJ5 A0A026WF57 A0A195DZB1 E9JCD1 A0A0J7L5G6 E2C6N6 A0A151XE67 E2A100 A0A1W4W5J5 A0A2J7RBA6 D6WMX7 A0A212F1M3 A0A154PPU2 A0A3L8DP12 A0A067QP10 A0A2A3E9N2 A0A088A8B6 A0A0N0BGS3 A0A1B0C851 A0A1Y1KNP7 A0A1L8E5A5 A0A0C9R7Z7 A0A336M2P4 A0A336KG51 A0A182K743 K7IRI4 A0A182UPG6 Q7QEC0 A0A182I9R1 A0A0L0C853 W8BVG6 V5GYU1 A0A182R3G4 A0A182MIH5 A0A034VTK6 A0A0A1XRA1 U5ET37 A0A182Y7J8 A0A182PPC7 A0A182Q473 T1DE60 U4U4A8 Q16RB5 A0A232EYU9 A0A0K8U9J5 A0A182W4B3 J3JTC6 A0A182MZP3 A0A1J1HP29 A0A1Q3F4A2 A0A1Q3F452 A0A1Q3F460 A0A1Q3EU60 A0A084VVR5 A0A1B6CNW6 W8B7V1 A0A1I8QAG0 A0A182J0Q9 A0A182UEL6 A0A023EUB2 T1PDU8 A0A182KY12 A0A182X575 B3MVY2 B4JJL4 B0WSX6 A0A0C9R4M3 Q9VZ20 A0A182H422 A0A3B0JWX1 A0A0R3P537 B4GXK5 B3NV95 A0A1W4V195 B4PYE4 A0A0K8TNC2 B4M2N2 A0A1B6HG70 A0A1B6F978 A0A224XG43 A0A1B6FUE1 A0A1A9WVD1 A0A0P4VTP7 B4L4H5 Q29HJ5 B4N2K3 A0A1B6ISY7 A0A069DUL0 A0A182S7P0 A0A0V0G5K0 A0A1W4UNU4 A0A1A9YP15 A0A1B0AXR0 A0A0Q9WKF4
Pubmed
26354079
22118469
21347285
21719571
24508170
21282665
+ More
20798317 18362917 19820115 30249741 24845553 28004739 20075255 12364791 14747013 17210077 26108605 24495485 25348373 25830018 25244985 24330624 23537049 17510324 28648823 22516182 24438588 24945155 25315136 20966253 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 15632085 17550304 26369729 27129103 23185243 26334808 18057021
20798317 18362917 19820115 30249741 24845553 28004739 20075255 12364791 14747013 17210077 26108605 24495485 25348373 25830018 25244985 24330624 23537049 17510324 28648823 22516182 24438588 24945155 25315136 20966253 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 15632085 17550304 26369729 27129103 23185243 26334808 18057021
EMBL
KQ460644
KPJ13223.1
KQ459584
KPI98532.1
AGBW02014067
OWR42327.1
+ More
KQ976465 KYM84270.1 KQ981820 KYN35281.1 ADTU01018485 ADTU01018486 ADTU01018487 GL888176 EGI65931.1 KK107276 EZA53634.1 KQ980031 KYN18161.1 GL771786 EFZ09519.1 LBMM01000500 KMQ98202.1 GL453161 EFN76348.1 KQ982254 KYQ58653.1 GL435626 EFN73053.1 NEVH01005920 PNF38108.1 KQ971343 EFA03267.1 AGBW02010853 OWR47632.1 KQ435022 KZC13915.1 QOIP01000006 RLU22026.1 KK853103 KDR11321.1 KZ288325 PBC27992.1 KQ435768 KOX75211.1 AJWK01000171 GEZM01078075 JAV63023.1 GFDF01000154 JAV13930.1 GBYB01002896 JAG72663.1 UFQT01000452 SSX24526.1 UFQS01000382 UFQT01000382 SSX03429.1 SSX23794.1 AAAB01008847 EAA06916.5 APCN01001027 JRES01000755 KNC28618.1 GAMC01013436 JAB93119.1 GALX01001424 JAB67042.1 AXCM01004867 GAKP01014069 JAC44883.1 GBXI01000951 GBXI01000741 JAD13341.1 JAD13551.1 GANO01002107 JAB57764.1 AXCN02001328 AXCN02001329 GALA01001177 JAA93675.1 KB632003 ERL87872.1 CH477719 EAT36932.1 NNAY01001561 OXU23594.1 GDHF01028977 JAI23337.1 APGK01044227 BT126482 KB741021 AEE61446.1 ENN75095.1 CVRI01000014 CRK89799.1 GFDL01012664 JAV22381.1 GFDL01012699 JAV22346.1 GFDL01012693 JAV22352.1 GFDL01016202 JAV18843.1 ATLV01017215 ATLV01017216 KE525157 KFB42059.1 GEDC01022180 JAS15118.1 GAMC01013437 JAB93118.1 GAPW01001092 JAC12506.1 KA646315 AFP60944.1 CH902625 EDV35127.2 CH916370 EDV99766.1 DS232077 EDS34104.1 GBYB01002895 JAG72662.1 AE014298 AY058302 AAF48011.2 AAL13531.1 AAN09289.1 JXUM01108602 KQ565246 KXJ71186.1 OUUW01000011 SPP86575.1 CH379064 KRT06512.1 CH479196 EDW27482.1 CH954180 EDV46083.1 CM000162 EDX02006.1 GDAI01001729 JAI15874.1 CH940651 EDW65936.1 GECU01033972 GECU01003756 JAS73734.1 JAT03951.1 GECZ01022972 JAS46797.1 GFTR01006422 JAW10004.1 GECZ01016166 JAS53603.1 GDKW01000589 JAI56006.1 CH933810 EDW07453.1 EAL31763.2 KRT06513.1 CH963925 EDW78592.1 GECU01017660 JAS90046.1 GBGD01001393 JAC87496.1 GECL01002780 JAP03344.1 JXJN01005350 KRF82487.1 KRF82488.1
KQ976465 KYM84270.1 KQ981820 KYN35281.1 ADTU01018485 ADTU01018486 ADTU01018487 GL888176 EGI65931.1 KK107276 EZA53634.1 KQ980031 KYN18161.1 GL771786 EFZ09519.1 LBMM01000500 KMQ98202.1 GL453161 EFN76348.1 KQ982254 KYQ58653.1 GL435626 EFN73053.1 NEVH01005920 PNF38108.1 KQ971343 EFA03267.1 AGBW02010853 OWR47632.1 KQ435022 KZC13915.1 QOIP01000006 RLU22026.1 KK853103 KDR11321.1 KZ288325 PBC27992.1 KQ435768 KOX75211.1 AJWK01000171 GEZM01078075 JAV63023.1 GFDF01000154 JAV13930.1 GBYB01002896 JAG72663.1 UFQT01000452 SSX24526.1 UFQS01000382 UFQT01000382 SSX03429.1 SSX23794.1 AAAB01008847 EAA06916.5 APCN01001027 JRES01000755 KNC28618.1 GAMC01013436 JAB93119.1 GALX01001424 JAB67042.1 AXCM01004867 GAKP01014069 JAC44883.1 GBXI01000951 GBXI01000741 JAD13341.1 JAD13551.1 GANO01002107 JAB57764.1 AXCN02001328 AXCN02001329 GALA01001177 JAA93675.1 KB632003 ERL87872.1 CH477719 EAT36932.1 NNAY01001561 OXU23594.1 GDHF01028977 JAI23337.1 APGK01044227 BT126482 KB741021 AEE61446.1 ENN75095.1 CVRI01000014 CRK89799.1 GFDL01012664 JAV22381.1 GFDL01012699 JAV22346.1 GFDL01012693 JAV22352.1 GFDL01016202 JAV18843.1 ATLV01017215 ATLV01017216 KE525157 KFB42059.1 GEDC01022180 JAS15118.1 GAMC01013437 JAB93118.1 GAPW01001092 JAC12506.1 KA646315 AFP60944.1 CH902625 EDV35127.2 CH916370 EDV99766.1 DS232077 EDS34104.1 GBYB01002895 JAG72662.1 AE014298 AY058302 AAF48011.2 AAL13531.1 AAN09289.1 JXUM01108602 KQ565246 KXJ71186.1 OUUW01000011 SPP86575.1 CH379064 KRT06512.1 CH479196 EDW27482.1 CH954180 EDV46083.1 CM000162 EDX02006.1 GDAI01001729 JAI15874.1 CH940651 EDW65936.1 GECU01033972 GECU01003756 JAS73734.1 JAT03951.1 GECZ01022972 JAS46797.1 GFTR01006422 JAW10004.1 GECZ01016166 JAS53603.1 GDKW01000589 JAI56006.1 CH933810 EDW07453.1 EAL31763.2 KRT06513.1 CH963925 EDW78592.1 GECU01017660 JAS90046.1 GBGD01001393 JAC87496.1 GECL01002780 JAP03344.1 JXJN01005350 KRF82487.1 KRF82488.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000078540
UP000078541
UP000005205
+ More
UP000007755 UP000053097 UP000078492 UP000036403 UP000008237 UP000075809 UP000000311 UP000192223 UP000235965 UP000007266 UP000076502 UP000279307 UP000027135 UP000242457 UP000005203 UP000053105 UP000092461 UP000075881 UP000002358 UP000075903 UP000007062 UP000075840 UP000037069 UP000075900 UP000075883 UP000076408 UP000075885 UP000075886 UP000030742 UP000008820 UP000215335 UP000075920 UP000019118 UP000075884 UP000183832 UP000030765 UP000095300 UP000075880 UP000075902 UP000095301 UP000075882 UP000076407 UP000007801 UP000001070 UP000002320 UP000000803 UP000069940 UP000249989 UP000268350 UP000001819 UP000008744 UP000008711 UP000192221 UP000002282 UP000008792 UP000091820 UP000009192 UP000007798 UP000075901 UP000092443 UP000092460
UP000007755 UP000053097 UP000078492 UP000036403 UP000008237 UP000075809 UP000000311 UP000192223 UP000235965 UP000007266 UP000076502 UP000279307 UP000027135 UP000242457 UP000005203 UP000053105 UP000092461 UP000075881 UP000002358 UP000075903 UP000007062 UP000075840 UP000037069 UP000075900 UP000075883 UP000076408 UP000075885 UP000075886 UP000030742 UP000008820 UP000215335 UP000075920 UP000019118 UP000075884 UP000183832 UP000030765 UP000095300 UP000075880 UP000075902 UP000095301 UP000075882 UP000076407 UP000007801 UP000001070 UP000002320 UP000000803 UP000069940 UP000249989 UP000268350 UP000001819 UP000008744 UP000008711 UP000192221 UP000002282 UP000008792 UP000091820 UP000009192 UP000007798 UP000075901 UP000092443 UP000092460
PRIDE
Interpro
ProteinModelPortal
A0A194R5X6
A0A194PYX3
A0A212ELH9
A0A195BHI9
A0A195F4H0
A0A158NJZ5
+ More
F4WIJ5 A0A026WF57 A0A195DZB1 E9JCD1 A0A0J7L5G6 E2C6N6 A0A151XE67 E2A100 A0A1W4W5J5 A0A2J7RBA6 D6WMX7 A0A212F1M3 A0A154PPU2 A0A3L8DP12 A0A067QP10 A0A2A3E9N2 A0A088A8B6 A0A0N0BGS3 A0A1B0C851 A0A1Y1KNP7 A0A1L8E5A5 A0A0C9R7Z7 A0A336M2P4 A0A336KG51 A0A182K743 K7IRI4 A0A182UPG6 Q7QEC0 A0A182I9R1 A0A0L0C853 W8BVG6 V5GYU1 A0A182R3G4 A0A182MIH5 A0A034VTK6 A0A0A1XRA1 U5ET37 A0A182Y7J8 A0A182PPC7 A0A182Q473 T1DE60 U4U4A8 Q16RB5 A0A232EYU9 A0A0K8U9J5 A0A182W4B3 J3JTC6 A0A182MZP3 A0A1J1HP29 A0A1Q3F4A2 A0A1Q3F452 A0A1Q3F460 A0A1Q3EU60 A0A084VVR5 A0A1B6CNW6 W8B7V1 A0A1I8QAG0 A0A182J0Q9 A0A182UEL6 A0A023EUB2 T1PDU8 A0A182KY12 A0A182X575 B3MVY2 B4JJL4 B0WSX6 A0A0C9R4M3 Q9VZ20 A0A182H422 A0A3B0JWX1 A0A0R3P537 B4GXK5 B3NV95 A0A1W4V195 B4PYE4 A0A0K8TNC2 B4M2N2 A0A1B6HG70 A0A1B6F978 A0A224XG43 A0A1B6FUE1 A0A1A9WVD1 A0A0P4VTP7 B4L4H5 Q29HJ5 B4N2K3 A0A1B6ISY7 A0A069DUL0 A0A182S7P0 A0A0V0G5K0 A0A1W4UNU4 A0A1A9YP15 A0A1B0AXR0 A0A0Q9WKF4
F4WIJ5 A0A026WF57 A0A195DZB1 E9JCD1 A0A0J7L5G6 E2C6N6 A0A151XE67 E2A100 A0A1W4W5J5 A0A2J7RBA6 D6WMX7 A0A212F1M3 A0A154PPU2 A0A3L8DP12 A0A067QP10 A0A2A3E9N2 A0A088A8B6 A0A0N0BGS3 A0A1B0C851 A0A1Y1KNP7 A0A1L8E5A5 A0A0C9R7Z7 A0A336M2P4 A0A336KG51 A0A182K743 K7IRI4 A0A182UPG6 Q7QEC0 A0A182I9R1 A0A0L0C853 W8BVG6 V5GYU1 A0A182R3G4 A0A182MIH5 A0A034VTK6 A0A0A1XRA1 U5ET37 A0A182Y7J8 A0A182PPC7 A0A182Q473 T1DE60 U4U4A8 Q16RB5 A0A232EYU9 A0A0K8U9J5 A0A182W4B3 J3JTC6 A0A182MZP3 A0A1J1HP29 A0A1Q3F4A2 A0A1Q3F452 A0A1Q3F460 A0A1Q3EU60 A0A084VVR5 A0A1B6CNW6 W8B7V1 A0A1I8QAG0 A0A182J0Q9 A0A182UEL6 A0A023EUB2 T1PDU8 A0A182KY12 A0A182X575 B3MVY2 B4JJL4 B0WSX6 A0A0C9R4M3 Q9VZ20 A0A182H422 A0A3B0JWX1 A0A0R3P537 B4GXK5 B3NV95 A0A1W4V195 B4PYE4 A0A0K8TNC2 B4M2N2 A0A1B6HG70 A0A1B6F978 A0A224XG43 A0A1B6FUE1 A0A1A9WVD1 A0A0P4VTP7 B4L4H5 Q29HJ5 B4N2K3 A0A1B6ISY7 A0A069DUL0 A0A182S7P0 A0A0V0G5K0 A0A1W4UNU4 A0A1A9YP15 A0A1B0AXR0 A0A0Q9WKF4
PDB
6F3A
E-value=6.93252e-108,
Score=999
Ontologies
GO
PANTHER
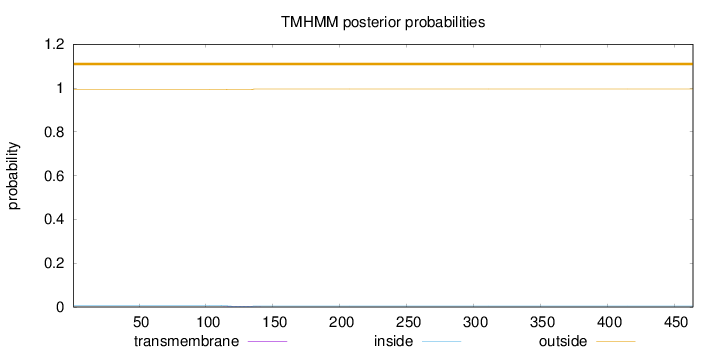
Topology
Length:
464
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0564300000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00615
outside
1 - 464
Population Genetic Test Statistics
Pi
228.603084
Theta
177.452441
Tajima's D
0.370305
CLR
1.174931
CSRT
0.47797610119494
Interpretation
Uncertain
