Gene
KWMTBOMO01707 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006026
Annotation
PREDICTED:_coiled-coil_and_C2_domain-containing_protein_1-like_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.187
Sequence
CDS
ATGTTTAGAAATCAGAAACCGGCAAAAAAGGGGAATCTGTCGCAGTTTGGTCTGTTAGATATTCCTGATTTGGACGATGCAGATGATGAACCACTAAATCTATCTGATGATGATATTGATTTGGAAGCAGAATTGGCTGCCATTAGTGGTGGCACAAGCAGACAGAGACCAAAAAAGGCTCCACCATCAGCCAATCTCGATGCCATGATAGCTGAAAGTTTAAAGGATATACCTTCAGATGAGGATGTTGATGAAGGCGACGAAGATGATCCAGACTTATTGAATGAATTAAAAAACTTAGGCCTCGATGATGAAGCTCCTCCAGCACCTCCGCCTCGTTCTTCCCGTCCTGCGCCTCCTCCTCCTGGAGCTGATACTGGCATTGTGGGGGTTTTACAGGACAGAATTAATAATTATATGGCTGCAGAAAAACATGCAAAGGAAAAGGGTGAAAGCAGTAGGGCTAGAAGATTCAACCGAGGTATAAAGACTCTAAATGATCTACTAAAGCAAGCTAAAGCTGGTAAACCCATCAGCGAAGAAGACATTCCACCAGCCGTAAGTCTTGGAAAACCTGCTGATTCCACACCATCAGCACCACTCTCTGGTCCAGTACAACCACCAACAGACCCAGAGCCACTACTGATAGATCTTGGACACGAGGATCCTCCCAGTTTACCAGAACCGCAAGAACCACCGCCCAGTCTACCGGAACCAGAGGAGCCACCACTGCCTCCACCGCGAGCTGAAAGCTTACACCAAGCAGACGATACCCTCACTCCTGAGAGACAGCAGAAGCTGCAGGCAATATTGAAACGTCAAGCAGAGTTTAAAGCGGCGGCGTTGCACTGTAAAAAATCGGGGGATAAAGCATTAGCACTCGAGTTTTTGAAGACAGTGAAGCAGTTCGATGTTCTGGTGACTGCTTTCAAAACGACCACCGAGGGGATGGACTTCGATTTAGCCGATTTACCTACTTTGGAGTCGGTCGCCGCGACCCTCAAGAGCCCAAAGGCCGATGAACAAATGCAAAATTCCGGAGAACCAACACCAGCGTCGACTCCAGCCTCCACGGAGCTGATCACCGCCTCGACGGTGGAAGAAGCTTTACGACAGAGGCTAGCTTATTTTCAGCAACAAGAGTCAAAAGCGAAGGAAGATGGTAACTCGTCGAAGGCGCGTCGCATGGGGCGCATTGTGAAGCAGTACCAGGAGGCGGTGCGGCTGCACGCGGCCGGGAGGGCGCTGCCCGGGGACGAGCTGCCCACGCCGCCGGGGTACGCGCCCCTACCCGCTGATCCAGCTGAAGCACCGCGCCCCGCCCCGCCCGCACCCCGCCCCGCGGCTCCGTCCCCCTCCCCTGCGCCGTCCCCCTCCGCTGCCCCGCGCCCCTCGTCCGCCCGCTACGAGAAGCAGCTCGCGTACCTGTTGATGCGCCAGAAACAGTTCAAGGAGGCCGCATTGAAAGCTAAAAAAGAAGGTAATATTGAACAAGCAAAAGAGTATTTACGTGCAGCCAAAGGGTTCGATACGGTGATAGACGCCGCGAGAGGCGGCCTGCCGGTCGACCTCAAGTCTTTGCCCCTCCCGCCTAAAGCTAAAGAGAATTTAGAACACACTTTCGACATAGTGTCGAGTGAGGACTGCGGCCCGTCGGACGAGGTGGCCACCGTGCGCGGCGAGGACGGCGACGTGTGGGCGCGCCTGCAGCAGCAGCTAGGCTCGCAGCTGCAGCTGTGCCAGCGGAACCGGGACCACTGCAGCGCCACCGGACACATCGCGGAGGCGAACAGGTTCGAGCACCTCGCCGTTACGGTCGCGCAGGACCTGGACGTCGTGGCGGTCGCCAAAAGCCTGGGCCGCGGTCCGCCGAAGTTCCACTACGAGTCCCGGCAGTTCGCGGTGGCGCAGTGCAACACGGACCTCAACGAGAACGAGCTGCAGCTGACCGTGGTCCGCGGCATCGCGTACAACGTCGCCAACCCGCAAGACGTCGACACTTACGTCAAATTCGAATTCCCCTACCCCCCGGAGGCGCCGGCGACGGACTGCACGGCGGTGGTGAAGGACACGAACAGCCCGGACTACGGCGCGGTGTTCTCGCTGCCCATCCAGCGCGCCGCCCGCACCTGCCAGCGCGCCTTCAAGCGGCACGCACTCAAGCTGCACGTGTACTCCAGGGGCGGCTGGTTCTCGCGGGACGCGCTGCTCGGGACCGTGGTCGTCCGCCTCGCGCCGCTGGAGACGCACGTCACCCTGCACGAGGCCTTCCCGCTGATGGACGGGCGCCGCCCTGCCGGTGGCTCGCTCGAGGTCAAACTGAGGGTCCGGACTCCGATCCTGCACCAGCAGATCGAAAACACGACGCATCGGTGGCTCGTCATCGATAACTAA
Protein
MFRNQKPAKKGNLSQFGLLDIPDLDDADDEPLNLSDDDIDLEAELAAISGGTSRQRPKKAPPSANLDAMIAESLKDIPSDEDVDEGDEDDPDLLNELKNLGLDDEAPPAPPPRSSRPAPPPPGADTGIVGVLQDRINNYMAAEKHAKEKGESSRARRFNRGIKTLNDLLKQAKAGKPISEEDIPPAVSLGKPADSTPSAPLSGPVQPPTDPEPLLIDLGHEDPPSLPEPQEPPPSLPEPEEPPLPPPRAESLHQADDTLTPERQQKLQAILKRQAEFKAAALHCKKSGDKALALEFLKTVKQFDVLVTAFKTTTEGMDFDLADLPTLESVAATLKSPKADEQMQNSGEPTPASTPASTELITASTVEEALRQRLAYFQQQESKAKEDGNSSKARRMGRIVKQYQEAVRLHAAGRALPGDELPTPPGYAPLPADPAEAPRPAPPAPRPAAPSPSPAPSPSAAPRPSSARYEKQLAYLLMRQKQFKEAALKAKKEGNIEQAKEYLRAAKGFDTVIDAARGGLPVDLKSLPLPPKAKENLEHTFDIVSSEDCGPSDEVATVRGEDGDVWARLQQQLGSQLQLCQRNRDHCSATGHIAEANRFEHLAVTVAQDLDVVAVAKSLGRGPPKFHYESRQFAVAQCNTDLNENELQLTVVRGIAYNVANPQDVDTYVKFEFPYPPEAPATDCTAVVKDTNSPDYGAVFSLPIQRAARTCQRAFKRHALKLHVYSRGGWFSRDALLGTVVVRLAPLETHVTLHEAFPLMDGRRPAGGSLEVKLRVRTPILHQQIENTTHRWLVIDN
Summary
Uniprot
H9J934
A0A194PYX9
A0A212ELF8
A0A2A4JVH4
D6WKC8
A0A2A4JV28
+ More
A0A1Y1LET1 A0A1Y1LA00 A0A2H1VKL3 A0A0T6B0P3 A0A1Y1LA18 A0A1Y1LA05 A0A2M4BDE7 A0A2M4AEX3 A0A2M4BDY6 A0A1Q3F686 A0A0L0CPN1 A0A182MYA9 A0A182K679 A0A182UCU7 Q17BV0 B0WWF2 W5JI35 A0A1Y1LFY4 A0A1Y1LC74 Q17BU9 A0A084WAV2 A0A1Q3F601 A0A182YF66 A0A1Q3F7A4 A0A1L8DB41 A0A1I8MMA6 T1PB41 A0A182P851 A0A1I8PLZ3 A0A0N1IT78 A0A182M0P2 A0A1I8MM95 A0A1I8PM59 A0A2R7X2H3 A0A195EBV3 F4WNH2 E2A806 A0A195FVZ6 A0A088A2G2 A0A1B0DAH3 A0A151IMA5 E9HDV2 A0A3L8E104 A0A158NTR3 A0A151WVK9 A0A195B7G7
A0A1Y1LET1 A0A1Y1LA00 A0A2H1VKL3 A0A0T6B0P3 A0A1Y1LA18 A0A1Y1LA05 A0A2M4BDE7 A0A2M4AEX3 A0A2M4BDY6 A0A1Q3F686 A0A0L0CPN1 A0A182MYA9 A0A182K679 A0A182UCU7 Q17BV0 B0WWF2 W5JI35 A0A1Y1LFY4 A0A1Y1LC74 Q17BU9 A0A084WAV2 A0A1Q3F601 A0A182YF66 A0A1Q3F7A4 A0A1L8DB41 A0A1I8MMA6 T1PB41 A0A182P851 A0A1I8PLZ3 A0A0N1IT78 A0A182M0P2 A0A1I8MM95 A0A1I8PM59 A0A2R7X2H3 A0A195EBV3 F4WNH2 E2A806 A0A195FVZ6 A0A088A2G2 A0A1B0DAH3 A0A151IMA5 E9HDV2 A0A3L8E104 A0A158NTR3 A0A151WVK9 A0A195B7G7
Pubmed
EMBL
BABH01004500
BABH01004501
BABH01004502
KQ459584
KPI98531.1
AGBW02014067
+ More
OWR42326.1 NWSH01000590 PCG75473.1 KQ971342 EFA03599.1 PCG75474.1 GEZM01061437 JAV70485.1 GEZM01061440 JAV70482.1 ODYU01003080 SOQ41389.1 LJIG01016420 KRT80654.1 GEZM01061438 JAV70484.1 GEZM01061439 JAV70483.1 GGFJ01001938 MBW51079.1 GGFK01006018 MBW39339.1 GGFJ01001887 MBW51028.1 GFDL01012022 JAV23023.1 JRES01000096 KNC34137.1 CH477316 EAT43762.1 DS232145 EDS36044.1 ADMH02001479 ETN62429.1 GEZM01061435 JAV70486.1 GEZM01061441 JAV70481.1 EAT43763.1 ATLV01022269 KE525331 KFB47346.1 GFDL01012063 JAV22982.1 GFDL01011600 JAV23445.1 GFDF01010489 JAV03595.1 KA645183 AFP59812.1 KQ435881 KOX69851.1 AXCM01000355 KK856478 PTY25861.1 KQ979074 KYN22705.1 GL888237 EGI64165.1 GL437468 EFN70443.1 KQ981215 KYN44601.1 AJVK01029070 AJVK01029071 AJVK01029072 KQ977063 KYN06003.1 GL732625 EFX70110.1 QOIP01000001 RLU26306.1 ADTU01002813 ADTU01002814 ADTU01002815 ADTU01002816 KQ982706 KYQ51846.1 KQ976565 KYM80456.1
OWR42326.1 NWSH01000590 PCG75473.1 KQ971342 EFA03599.1 PCG75474.1 GEZM01061437 JAV70485.1 GEZM01061440 JAV70482.1 ODYU01003080 SOQ41389.1 LJIG01016420 KRT80654.1 GEZM01061438 JAV70484.1 GEZM01061439 JAV70483.1 GGFJ01001938 MBW51079.1 GGFK01006018 MBW39339.1 GGFJ01001887 MBW51028.1 GFDL01012022 JAV23023.1 JRES01000096 KNC34137.1 CH477316 EAT43762.1 DS232145 EDS36044.1 ADMH02001479 ETN62429.1 GEZM01061435 JAV70486.1 GEZM01061441 JAV70481.1 EAT43763.1 ATLV01022269 KE525331 KFB47346.1 GFDL01012063 JAV22982.1 GFDL01011600 JAV23445.1 GFDF01010489 JAV03595.1 KA645183 AFP59812.1 KQ435881 KOX69851.1 AXCM01000355 KK856478 PTY25861.1 KQ979074 KYN22705.1 GL888237 EGI64165.1 GL437468 EFN70443.1 KQ981215 KYN44601.1 AJVK01029070 AJVK01029071 AJVK01029072 KQ977063 KYN06003.1 GL732625 EFX70110.1 QOIP01000001 RLU26306.1 ADTU01002813 ADTU01002814 ADTU01002815 ADTU01002816 KQ982706 KYQ51846.1 KQ976565 KYM80456.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000218220
UP000007266
UP000037069
+ More
UP000075884 UP000075881 UP000075902 UP000008820 UP000002320 UP000000673 UP000030765 UP000076408 UP000095301 UP000075885 UP000095300 UP000053105 UP000075883 UP000078492 UP000007755 UP000000311 UP000078541 UP000005203 UP000092462 UP000078542 UP000000305 UP000279307 UP000005205 UP000075809 UP000078540
UP000075884 UP000075881 UP000075902 UP000008820 UP000002320 UP000000673 UP000030765 UP000076408 UP000095301 UP000075885 UP000095300 UP000053105 UP000075883 UP000078492 UP000007755 UP000000311 UP000078541 UP000005203 UP000092462 UP000078542 UP000000305 UP000279307 UP000005205 UP000075809 UP000078540
PRIDE
Pfam
PF00168 C2
Interpro
Gene 3D
ProteinModelPortal
H9J934
A0A194PYX9
A0A212ELF8
A0A2A4JVH4
D6WKC8
A0A2A4JV28
+ More
A0A1Y1LET1 A0A1Y1LA00 A0A2H1VKL3 A0A0T6B0P3 A0A1Y1LA18 A0A1Y1LA05 A0A2M4BDE7 A0A2M4AEX3 A0A2M4BDY6 A0A1Q3F686 A0A0L0CPN1 A0A182MYA9 A0A182K679 A0A182UCU7 Q17BV0 B0WWF2 W5JI35 A0A1Y1LFY4 A0A1Y1LC74 Q17BU9 A0A084WAV2 A0A1Q3F601 A0A182YF66 A0A1Q3F7A4 A0A1L8DB41 A0A1I8MMA6 T1PB41 A0A182P851 A0A1I8PLZ3 A0A0N1IT78 A0A182M0P2 A0A1I8MM95 A0A1I8PM59 A0A2R7X2H3 A0A195EBV3 F4WNH2 E2A806 A0A195FVZ6 A0A088A2G2 A0A1B0DAH3 A0A151IMA5 E9HDV2 A0A3L8E104 A0A158NTR3 A0A151WVK9 A0A195B7G7
A0A1Y1LET1 A0A1Y1LA00 A0A2H1VKL3 A0A0T6B0P3 A0A1Y1LA18 A0A1Y1LA05 A0A2M4BDE7 A0A2M4AEX3 A0A2M4BDY6 A0A1Q3F686 A0A0L0CPN1 A0A182MYA9 A0A182K679 A0A182UCU7 Q17BV0 B0WWF2 W5JI35 A0A1Y1LFY4 A0A1Y1LC74 Q17BU9 A0A084WAV2 A0A1Q3F601 A0A182YF66 A0A1Q3F7A4 A0A1L8DB41 A0A1I8MMA6 T1PB41 A0A182P851 A0A1I8PLZ3 A0A0N1IT78 A0A182M0P2 A0A1I8MM95 A0A1I8PM59 A0A2R7X2H3 A0A195EBV3 F4WNH2 E2A806 A0A195FVZ6 A0A088A2G2 A0A1B0DAH3 A0A151IMA5 E9HDV2 A0A3L8E104 A0A158NTR3 A0A151WVK9 A0A195B7G7
PDB
6EI6
E-value=1.99721e-72,
Score=696
Ontologies
GO
PANTHER
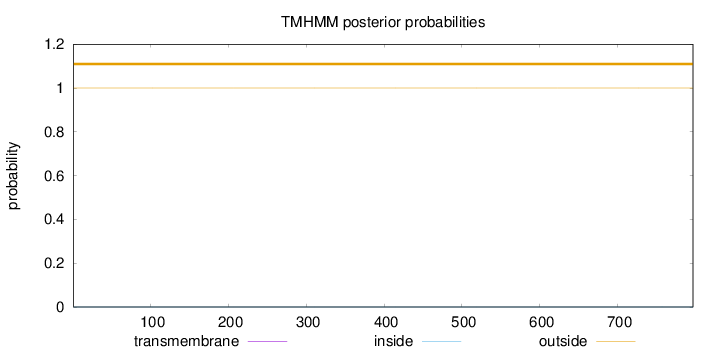
Topology
Length:
797
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0016
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00028
outside
1 - 797
Population Genetic Test Statistics
Pi
193.103408
Theta
176.945452
Tajima's D
0.382483
CLR
0.386323
CSRT
0.483975801209939
Interpretation
Uncertain
