Gene
KWMTBOMO01697
Pre Gene Modal
BGIBMGA006117
Annotation
PREDICTED:_tripartite_motif-containing_protein_45_[Bombyx_mori]
Full name
Tripartite motif-containing protein 45
Alternative Name
RING finger protein 99
Location in the cell
Nuclear Reliability : 2.8
Sequence
CDS
ATGAATAATTTATGCTTCACCCGCAAGAAGGACAAGCGGAAGTCACTGGAGCCCTCATCAATAAGTGCCGGTAATAGTCCTTTAAGGACCCGACCGGCTAGCCAAAGCCTGTCTATAGCGCCAAGGACCTCAGCTTACGACAAAAAAGATGTAATGAAGGAGTGGATCTGCGGGATATGTAGGAAAGAATTGGTGGAACCGAGGCTGCTGGGCTGCCTCCACAGCTTCTGCACCAGCTGTCTTCAGGGTCTGCATCAGGAGGGTGCTTGTGAATTGTGGAGCGAAGTCGATAGAGAGTCAATCCAACAAGATCCTGCCGAATCAGGATCAGGCTTGGGTTCCGCCGGATCCGGATACGAGAGTGACTTGAGACATTCAGACTCTGAGGGCAGTTGGGAGCACAAAAACAAGAAATACGGTATTCTGACAAGAAAGATTTCCGGTAAAAGCGTGCAATTCGTAGTATGCCCAACATGCGGTCAAGAGAGCGCGCTCCCTCTGGGAGGTATAGCGGCGTTGCCCCTCAACTACGTGCTGCTTCGGAAGATGAACTCACATGAGAACGGTGTGGCGGTACTCTGCGACCTTTGCGACAGTGACCATAAGGCTGTGAGTCGTTGCGGGAAATGCCTGATCAGCATATGCTCATCGTGCGGCGACGCCCACGAGAGGCAGCCGACCACGGCGAGACACGTACTGGAGCCGCTACACCCTCCCGTCAGATACTGCTCGCAACATCCCAAGGCTGAACTGTGCATCTACTGCGCTACTTGTCAACAGGTTGTGTGCCGAGAGTGCTGCGTGGTGTCGCACGGCGGGCACGCGCTGGCGGCGGCGTCCCGCGCGGCTGCCGAGCGCGCCCGCGTGCTGCGTGCCGCCTGCGATCGCGCCGCGCACGTGCCGGGCAACGTGCAGCGGGCGGCCAGGGCGCTGCAAGTGCATGCCGCTCAAGTTGATGCTCAGGCTTCCAAAGTGGAATCGGAAGTGGAGACGTGGGCGGCGGAGTACAGTCGCGCGGTGGAGGCGCACGGGCGCGCGCTGCTGCGGGCCGCGGGGCGGGCGCGCTGCGGGTTCCGTCGCGGCGTGCGGCGCCACGAGCAGCGCCTCGACGAGCGCGTGCGGGACGCGCACGAAGCCGTCGCCTTCGCCCAGGAGTTGCTCTCGGGAGCCAGAGACGATGAGTTGCTATCCCTGAGCGGGCCCGTCCTCCGGCGGTTAGAGAGGGTGACGGAGGTAGAGGGGCTGGGGCAGGCTCCCCAGTGCGAGGTCAGGTTCGCGCCGCTGGCCCCCGCCGCGCCGCACTCCACGCTGTACGGGCGGCTGCTCACTCAGGCGCCGGACCCCGACAAATGCGTTCTGGATACTGAAGGTCTCCAAGACCTGATGGTGAATTGTCAACACGAGACAGTTTTAGAGCTGAGGGACTGTAACGGTGAGCGCATTTGGTGCGGAGGCGAACAGGTGTCGGGCTACCTGCGGCGCCGCGACTCGAGCGCGCGGCCGGCCGGCGCCGCGGTTACGGACCGCGCCGACGGTCGCTACACGCTCGCCTTCACGCCCGCCGCGCCCGGCGCATACCTGCTGGCCGTCACCGTCGCCAACAAGCCTATTAAGGTAACACCGACACCGGTCCGGTCCGGTCACCACTCTCGTCGAGCCCGTCGCTTGCGACATAGGGCTCGGTGA
Protein
MNNLCFTRKKDKRKSLEPSSISAGNSPLRTRPASQSLSIAPRTSAYDKKDVMKEWICGICRKELVEPRLLGCLHSFCTSCLQGLHQEGACELWSEVDRESIQQDPAESGSGLGSAGSGYESDLRHSDSEGSWEHKNKKYGILTRKISGKSVQFVVCPTCGQESALPLGGIAALPLNYVLLRKMNSHENGVAVLCDLCDSDHKAVSRCGKCLISICSSCGDAHERQPTTARHVLEPLHPPVRYCSQHPKAELCIYCATCQQVVCRECCVVSHGGHALAAASRAAAERARVLRAACDRAAHVPGNVQRAARALQVHAAQVDAQASKVESEVETWAAEYSRAVEAHGRALLRAAGRARCGFRRGVRRHEQRLDERVRDAHEAVAFAQELLSGARDDELLSLSGPVLRRLERVTEVEGLGQAPQCEVRFAPLAPAAPHSTLYGRLLTQAPDPDKCVLDTEGLQDLMVNCQHETVLELRDCNGERIWCGGEQVSGYLRRRDSSARPAGAAVTDRADGRYTLAFTPAAPGAYLLAVTVANKPIKVTPTPVRSGHHSRRARRLRHRAR
Summary
Description
May act as a transcriptional repressor in mitogen-activated protein kinase signaling pathway.
Similarity
Belongs to the peptidase S1 family.
Belongs to the TRIM/RBCC family.
Belongs to the TRIM/RBCC family.
Keywords
Coiled coil
Complete proteome
Cytoplasm
Metal-binding
Nucleus
Reference proteome
Repeat
Zinc
Zinc-finger
3D-structure
Alternative splicing
Polymorphism
Feature
chain Tripartite motif-containing protein 45
splice variant In isoform 2.
sequence variant In dbSNP:rs34863850.
splice variant In isoform 2.
sequence variant In dbSNP:rs34863850.
Uniprot
H9J9C5
A0A2W1BH59
A0A2H1W5L3
A0A212EUM6
A0A194Q0K3
A0A194R7L5
+ More
A0A2A4JDV3 A0A0L7LI21 A0A067QS44 A0A1Y1NGD9 A0A2J7PUX6 A0A139WGX8 A0A1W4XKZ8 N6U9I2 U4UGS1 A0A146LE64 A0A336M470 A0A0A1WSU3 A0A0L0BV24 A0A034WHX8 K7GAL8 A0A0K8TZ76 A0A0K8WL26 W8BND0 B4G921 Q29ML5 B3N718 E0VVI1 B4NWF1 Q3ZAM2 Q9VLR7 B4Q6B4 B3MPI3 B4HYG2 A0A1W4UV96 T1GU90 A0A1I8PK99 B4MVX4 W5JIX1 A0A250YJX4 D2HMJ2 U3E940 A0A2Y9IV36 A0A2K5PDD8 G1QZH5 A0A2K6PGQ7 U6CTY3 G9KV41 G5BFJ1 I3L6F2 G3SJ16 A0A2K5KHR7 H0WUL0 A0A2K5YZ01 E2RGD5 K7GR26 A0A2K6CHA8 A0A0D9S639 G7NX54 G7MFZ7 C3Z575 A0A384DNI2 I3M9K9 M3W3N1 K7GLN7 L8YCK3 W5QGE2 K9IMF5 A0A1B0ANZ9 G3H907 A0A3L7IFW3 F7E8F4 A0A2K5IF96 B4KGW7 S7MIH9 D4A7S9 A0A2U3Z157 A0A2I3TLJ8 K7BMQ7 A0A1U7QX26 A0A383Z2R1 L8I6N5 Q5BIM1 A0A2J8SQM0 A0A2Y9P0D6 A0A2U3V5L6 H2N678 K7D444 K7AS39 A0A2K5C9J5 Q9H8W5 W5MZ37 A0A096NE79 A0A212CHA0 A0A2Y9H995 A0A341D0N3 F6RUP9 L5LXM3 A0A182H059 G3X1J0
A0A2A4JDV3 A0A0L7LI21 A0A067QS44 A0A1Y1NGD9 A0A2J7PUX6 A0A139WGX8 A0A1W4XKZ8 N6U9I2 U4UGS1 A0A146LE64 A0A336M470 A0A0A1WSU3 A0A0L0BV24 A0A034WHX8 K7GAL8 A0A0K8TZ76 A0A0K8WL26 W8BND0 B4G921 Q29ML5 B3N718 E0VVI1 B4NWF1 Q3ZAM2 Q9VLR7 B4Q6B4 B3MPI3 B4HYG2 A0A1W4UV96 T1GU90 A0A1I8PK99 B4MVX4 W5JIX1 A0A250YJX4 D2HMJ2 U3E940 A0A2Y9IV36 A0A2K5PDD8 G1QZH5 A0A2K6PGQ7 U6CTY3 G9KV41 G5BFJ1 I3L6F2 G3SJ16 A0A2K5KHR7 H0WUL0 A0A2K5YZ01 E2RGD5 K7GR26 A0A2K6CHA8 A0A0D9S639 G7NX54 G7MFZ7 C3Z575 A0A384DNI2 I3M9K9 M3W3N1 K7GLN7 L8YCK3 W5QGE2 K9IMF5 A0A1B0ANZ9 G3H907 A0A3L7IFW3 F7E8F4 A0A2K5IF96 B4KGW7 S7MIH9 D4A7S9 A0A2U3Z157 A0A2I3TLJ8 K7BMQ7 A0A1U7QX26 A0A383Z2R1 L8I6N5 Q5BIM1 A0A2J8SQM0 A0A2Y9P0D6 A0A2U3V5L6 H2N678 K7D444 K7AS39 A0A2K5C9J5 Q9H8W5 W5MZ37 A0A096NE79 A0A212CHA0 A0A2Y9H995 A0A341D0N3 F6RUP9 L5LXM3 A0A182H059 G3X1J0
Pubmed
19121390
28756777
22118469
26354079
26227816
24845553
+ More
28004739 18362917 19820115 23537049 26823975 25830018 26108605 25348373 17381049 24495485 17994087 15632085 20566863 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20920257 23761445 28087693 20010809 25243066 25362486 23236062 21993625 30723633 22398555 16341006 22002653 25319552 18563158 24813606 17975172 23385571 20809919 21804562 29704459 17495919 15057822 15632090 16136131 22751099 16305752 15351693 14702039 16710414 15489334 19892987 26483478 21709235
28004739 18362917 19820115 23537049 26823975 25830018 26108605 25348373 17381049 24495485 17994087 15632085 20566863 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20920257 23761445 28087693 20010809 25243066 25362486 23236062 21993625 30723633 22398555 16341006 22002653 25319552 18563158 24813606 17975172 23385571 20809919 21804562 29704459 17495919 15057822 15632090 16136131 22751099 16305752 15351693 14702039 16710414 15489334 19892987 26483478 21709235
EMBL
BABH01004468
BABH01004469
BABH01004470
KZ150052
PZC74382.1
ODYU01006354
+ More
SOQ48132.1 AGBW02012334 OWR45195.1 KQ459584 KPI98524.1 KQ460644 KPJ13230.1 NWSH01001783 PCG70151.1 JTDY01001103 KOB74876.1 KK853366 KDR08100.1 GEZM01003004 GEZM01003003 JAV96994.1 NEVH01021189 PNF20133.1 KQ971343 KYB27222.1 APGK01037333 APGK01037334 KB740948 ENN77306.1 KB632288 ERL91548.1 GDHC01012710 JAQ05919.1 UFQS01000320 UFQT01000320 SSX02829.1 SSX23197.1 GBXI01012717 JAD01575.1 JRES01001418 KNC23094.1 GAKP01005227 JAC53725.1 AGCU01152279 GDHF01032723 JAI19591.1 GDHF01000542 JAI51772.1 GAMC01006228 JAC00328.1 CH479180 EDW28851.1 CH379060 EAL33678.2 CH954177 EDV59314.1 AAZO01005641 DS235812 EEB17387.1 CM000157 EDW88468.1 BT023847 AAZ86768.1 AE014134 BT081973 AAF52617.1 ACO53088.1 CM000361 CM002910 EDX04190.1 KMY88975.1 CH902620 EDV31279.1 CH480818 EDW52092.1 CAQQ02017106 CAQQ02017107 CH963857 EDW75844.2 ADMH02001111 ETN64076.1 GFFW01000865 JAV43923.1 ACTA01130516 GL193045 EFB13582.1 GAMT01006772 GAMS01004671 JAB05089.1 JAB18465.1 ADFV01041685 ADFV01041686 HAAF01002339 CCP74165.1 JP020172 AES08770.1 JH170044 GEBF01005209 EHB08052.1 JAN98423.1 AEMK02000024 DQIR01187376 HDB42853.1 CABD030005566 AAQR03105779 AAQR03105780 AAQR03105781 AAEX03011013 DQIR01100779 DQIR01256175 DQIR01259368 DQIR01259369 DQIR01264626 DQIR01273752 DQIR01318016 HDA56255.1 HDC11653.1 AQIB01086232 AQIA01002779 CM001276 EHH50207.1 JU336496 JV045067 CM001253 AFE80249.1 AFI35138.1 EHH15100.1 GG666582 EEN52432.1 AGTP01080053 AANG04003279 DQIR01306602 HDC62080.1 KB359359 ELV14148.1 AMGL01004359 GABZ01005218 JAA48307.1 JXJN01001093 JXJN01001094 JXJN01001095 JH000221 EGW05805.1 RAZU01000058 RLQ76985.1 CH933807 EDW11167.1 KE161437 EPQ03771.1 AABR07012715 CH474015 EDL85532.1 AACZ04034370 GABC01003802 JAA07536.1 GABD01001549 NBAG03000423 JAA31551.1 PNI26405.1 JH881870 ELR51863.1 BT021203 NDHI03003555 PNJ23052.1 ABGA01029953 GABE01010220 JAA34519.1 GABF01001389 JAA20756.1 AY669488 AK023243 AK222901 AL391476 BC034943 AHAT01003684 AHAT01003685 AHZZ02016712 AHZZ02016713 MKHE01000020 OWK05423.1 KB106585 ELK30851.1 JXUM01100705 JXUM01100706 KQ564591 KXJ72048.1 AEFK01176495
SOQ48132.1 AGBW02012334 OWR45195.1 KQ459584 KPI98524.1 KQ460644 KPJ13230.1 NWSH01001783 PCG70151.1 JTDY01001103 KOB74876.1 KK853366 KDR08100.1 GEZM01003004 GEZM01003003 JAV96994.1 NEVH01021189 PNF20133.1 KQ971343 KYB27222.1 APGK01037333 APGK01037334 KB740948 ENN77306.1 KB632288 ERL91548.1 GDHC01012710 JAQ05919.1 UFQS01000320 UFQT01000320 SSX02829.1 SSX23197.1 GBXI01012717 JAD01575.1 JRES01001418 KNC23094.1 GAKP01005227 JAC53725.1 AGCU01152279 GDHF01032723 JAI19591.1 GDHF01000542 JAI51772.1 GAMC01006228 JAC00328.1 CH479180 EDW28851.1 CH379060 EAL33678.2 CH954177 EDV59314.1 AAZO01005641 DS235812 EEB17387.1 CM000157 EDW88468.1 BT023847 AAZ86768.1 AE014134 BT081973 AAF52617.1 ACO53088.1 CM000361 CM002910 EDX04190.1 KMY88975.1 CH902620 EDV31279.1 CH480818 EDW52092.1 CAQQ02017106 CAQQ02017107 CH963857 EDW75844.2 ADMH02001111 ETN64076.1 GFFW01000865 JAV43923.1 ACTA01130516 GL193045 EFB13582.1 GAMT01006772 GAMS01004671 JAB05089.1 JAB18465.1 ADFV01041685 ADFV01041686 HAAF01002339 CCP74165.1 JP020172 AES08770.1 JH170044 GEBF01005209 EHB08052.1 JAN98423.1 AEMK02000024 DQIR01187376 HDB42853.1 CABD030005566 AAQR03105779 AAQR03105780 AAQR03105781 AAEX03011013 DQIR01100779 DQIR01256175 DQIR01259368 DQIR01259369 DQIR01264626 DQIR01273752 DQIR01318016 HDA56255.1 HDC11653.1 AQIB01086232 AQIA01002779 CM001276 EHH50207.1 JU336496 JV045067 CM001253 AFE80249.1 AFI35138.1 EHH15100.1 GG666582 EEN52432.1 AGTP01080053 AANG04003279 DQIR01306602 HDC62080.1 KB359359 ELV14148.1 AMGL01004359 GABZ01005218 JAA48307.1 JXJN01001093 JXJN01001094 JXJN01001095 JH000221 EGW05805.1 RAZU01000058 RLQ76985.1 CH933807 EDW11167.1 KE161437 EPQ03771.1 AABR07012715 CH474015 EDL85532.1 AACZ04034370 GABC01003802 JAA07536.1 GABD01001549 NBAG03000423 JAA31551.1 PNI26405.1 JH881870 ELR51863.1 BT021203 NDHI03003555 PNJ23052.1 ABGA01029953 GABE01010220 JAA34519.1 GABF01001389 JAA20756.1 AY669488 AK023243 AK222901 AL391476 BC034943 AHAT01003684 AHAT01003685 AHZZ02016712 AHZZ02016713 MKHE01000020 OWK05423.1 KB106585 ELK30851.1 JXUM01100705 JXUM01100706 KQ564591 KXJ72048.1 AEFK01176495
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000037510
+ More
UP000027135 UP000235965 UP000007266 UP000192223 UP000019118 UP000030742 UP000037069 UP000007267 UP000008744 UP000001819 UP000008711 UP000009046 UP000002282 UP000000803 UP000000304 UP000007801 UP000001292 UP000192221 UP000015102 UP000095300 UP000007798 UP000000673 UP000008912 UP000008225 UP000248482 UP000233040 UP000001073 UP000233200 UP000006813 UP000008227 UP000001519 UP000233060 UP000005225 UP000233140 UP000002254 UP000233120 UP000029965 UP000009130 UP000233100 UP000001554 UP000261680 UP000291021 UP000005215 UP000011712 UP000011518 UP000002356 UP000092460 UP000001075 UP000273346 UP000002280 UP000233080 UP000009192 UP000002494 UP000245341 UP000002277 UP000189706 UP000261681 UP000009136 UP000248483 UP000245320 UP000001595 UP000233020 UP000005640 UP000018468 UP000028761 UP000248481 UP000252040 UP000002281 UP000069940 UP000249989 UP000007648
UP000027135 UP000235965 UP000007266 UP000192223 UP000019118 UP000030742 UP000037069 UP000007267 UP000008744 UP000001819 UP000008711 UP000009046 UP000002282 UP000000803 UP000000304 UP000007801 UP000001292 UP000192221 UP000015102 UP000095300 UP000007798 UP000000673 UP000008912 UP000008225 UP000248482 UP000233040 UP000001073 UP000233200 UP000006813 UP000008227 UP000001519 UP000233060 UP000005225 UP000233140 UP000002254 UP000233120 UP000029965 UP000009130 UP000233100 UP000001554 UP000261680 UP000291021 UP000005215 UP000011712 UP000011518 UP000002356 UP000092460 UP000001075 UP000273346 UP000002280 UP000233080 UP000009192 UP000002494 UP000245341 UP000002277 UP000189706 UP000261681 UP000009136 UP000248483 UP000245320 UP000001595 UP000233020 UP000005640 UP000018468 UP000028761 UP000248481 UP000252040 UP000002281 UP000069940 UP000249989 UP000007648
Interpro
IPR001841
Znf_RING
+ More
IPR027370 Znf-RING_LisH
IPR017907 Znf_RING_CS
IPR013783 Ig-like_fold
IPR001298 Filamin/ABP280_rpt
IPR014756 Ig_E-set
IPR000315 Znf_B-box
IPR017868 Filamin/ABP280_repeat-like
IPR018957 Znf_C3HC4_RING-type
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR022700 CLIP
IPR033116 TRYPSIN_SER
IPR013083 Znf_RING/FYVE/PHD
IPR003649 Bbox_C
IPR027370 Znf-RING_LisH
IPR017907 Znf_RING_CS
IPR013783 Ig-like_fold
IPR001298 Filamin/ABP280_rpt
IPR014756 Ig_E-set
IPR000315 Znf_B-box
IPR017868 Filamin/ABP280_repeat-like
IPR018957 Znf_C3HC4_RING-type
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR022700 CLIP
IPR033116 TRYPSIN_SER
IPR013083 Znf_RING/FYVE/PHD
IPR003649 Bbox_C
Gene 3D
ProteinModelPortal
H9J9C5
A0A2W1BH59
A0A2H1W5L3
A0A212EUM6
A0A194Q0K3
A0A194R7L5
+ More
A0A2A4JDV3 A0A0L7LI21 A0A067QS44 A0A1Y1NGD9 A0A2J7PUX6 A0A139WGX8 A0A1W4XKZ8 N6U9I2 U4UGS1 A0A146LE64 A0A336M470 A0A0A1WSU3 A0A0L0BV24 A0A034WHX8 K7GAL8 A0A0K8TZ76 A0A0K8WL26 W8BND0 B4G921 Q29ML5 B3N718 E0VVI1 B4NWF1 Q3ZAM2 Q9VLR7 B4Q6B4 B3MPI3 B4HYG2 A0A1W4UV96 T1GU90 A0A1I8PK99 B4MVX4 W5JIX1 A0A250YJX4 D2HMJ2 U3E940 A0A2Y9IV36 A0A2K5PDD8 G1QZH5 A0A2K6PGQ7 U6CTY3 G9KV41 G5BFJ1 I3L6F2 G3SJ16 A0A2K5KHR7 H0WUL0 A0A2K5YZ01 E2RGD5 K7GR26 A0A2K6CHA8 A0A0D9S639 G7NX54 G7MFZ7 C3Z575 A0A384DNI2 I3M9K9 M3W3N1 K7GLN7 L8YCK3 W5QGE2 K9IMF5 A0A1B0ANZ9 G3H907 A0A3L7IFW3 F7E8F4 A0A2K5IF96 B4KGW7 S7MIH9 D4A7S9 A0A2U3Z157 A0A2I3TLJ8 K7BMQ7 A0A1U7QX26 A0A383Z2R1 L8I6N5 Q5BIM1 A0A2J8SQM0 A0A2Y9P0D6 A0A2U3V5L6 H2N678 K7D444 K7AS39 A0A2K5C9J5 Q9H8W5 W5MZ37 A0A096NE79 A0A212CHA0 A0A2Y9H995 A0A341D0N3 F6RUP9 L5LXM3 A0A182H059 G3X1J0
A0A2A4JDV3 A0A0L7LI21 A0A067QS44 A0A1Y1NGD9 A0A2J7PUX6 A0A139WGX8 A0A1W4XKZ8 N6U9I2 U4UGS1 A0A146LE64 A0A336M470 A0A0A1WSU3 A0A0L0BV24 A0A034WHX8 K7GAL8 A0A0K8TZ76 A0A0K8WL26 W8BND0 B4G921 Q29ML5 B3N718 E0VVI1 B4NWF1 Q3ZAM2 Q9VLR7 B4Q6B4 B3MPI3 B4HYG2 A0A1W4UV96 T1GU90 A0A1I8PK99 B4MVX4 W5JIX1 A0A250YJX4 D2HMJ2 U3E940 A0A2Y9IV36 A0A2K5PDD8 G1QZH5 A0A2K6PGQ7 U6CTY3 G9KV41 G5BFJ1 I3L6F2 G3SJ16 A0A2K5KHR7 H0WUL0 A0A2K5YZ01 E2RGD5 K7GR26 A0A2K6CHA8 A0A0D9S639 G7NX54 G7MFZ7 C3Z575 A0A384DNI2 I3M9K9 M3W3N1 K7GLN7 L8YCK3 W5QGE2 K9IMF5 A0A1B0ANZ9 G3H907 A0A3L7IFW3 F7E8F4 A0A2K5IF96 B4KGW7 S7MIH9 D4A7S9 A0A2U3Z157 A0A2I3TLJ8 K7BMQ7 A0A1U7QX26 A0A383Z2R1 L8I6N5 Q5BIM1 A0A2J8SQM0 A0A2Y9P0D6 A0A2U3V5L6 H2N678 K7D444 K7AS39 A0A2K5C9J5 Q9H8W5 W5MZ37 A0A096NE79 A0A212CHA0 A0A2Y9H995 A0A341D0N3 F6RUP9 L5LXM3 A0A182H059 G3X1J0
PDB
2DS4
E-value=6.40868e-06,
Score=121
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
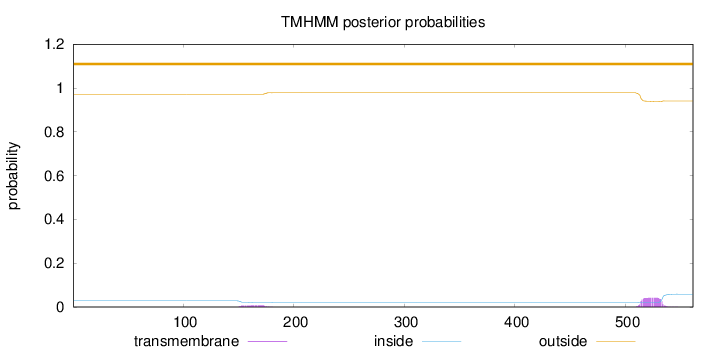
Length:
561
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.01146
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02809
outside
1 - 561
Population Genetic Test Statistics
Pi
156.495806
Theta
146.039187
Tajima's D
0.385543
CLR
0.461393
CSRT
0.482125893705315
Interpretation
Uncertain
