Gene
KWMTBOMO01696
Pre Gene Modal
BGIBMGA006029
Annotation
PREDICTED:_heparin_sulfate_O-sulfotransferase_[Amyelois_transitella]
Full name
Heparin sulfate O-sulfotransferase
Location in the cell
Nuclear Reliability : 2.128
Sequence
CDS
ATGGAAGAGAATTATCGTAAATTAGAAATAAGATTAGCCCACCCAGGAATCGGATTAACGGAGTCTCGCCAGTTTCCTCTAAATAATGCGGACCAGACTGTCATTATATACAATAGGGTCCCGAAGACGGGGTCCACGAGCTTCGTGGGAATCGCGTACGATTTATGCAAGAGGAACAATTTTAAAGTGCTACATATCAACATTACCGCCAACAGACATATCATGTCGTTGAATAACCAATATAAATTTGCTCAGAATGTAACCACTTGGCATGATGTCAAACCTGCCCTATATCATGGGCATATGGCCTTCCTAAATTTTGAGAGATTAGGTACATCATCAAAACCCCTATTCATAAATTTAATAAGGAAACCATTGGACAGACTAGTTTCATATTATTACTTCCTACGCTACGGTGACAATTTCCGACCCCATTTGGTACGAAAGAAACATGGTGACAAAATGACATTTGATGAATGTGTTGAGAAGAGCCAGCCGGATTGTGACCCAAACAATATGTGGTTACAAGTGCCTTTTTTCTGTGGTCACGCTGCAGAGTGCTGGAAACCAGGCAACCATTGGGCTTTAGAACAAGCAAAGCACAACTTAGTCAACCACTACCTTTTGGTAGGAGTCACCGAGGAAATGCAGGACTTCATCACAGTACTCGAAGCCACTTTACCGCGCTTCTTCAAAGGGGCCACCGAGTATTACCTAAGCAGTAATAAGTCCCACCTACGGCAGACGAGCTCGAAAGTGGACCCGAGCCTGAAGACGGTCGAGAGGATTCAACAGTCGACCGTTTGGAAAATGGAAAATGAGCTTTATGAGTTTGCATTAGAACATTTTAAATTTGTGAAAAGGAAACTTTTAGTGAAGGAAGCAAACAATGTGGCTCAGATATATTTTTATGAGAAAATAAGACCTAAATGA
Protein
MEENYRKLEIRLAHPGIGLTESRQFPLNNADQTVIIYNRVPKTGSTSFVGIAYDLCKRNNFKVLHINITANRHIMSLNNQYKFAQNVTTWHDVKPALYHGHMAFLNFERLGTSSKPLFINLIRKPLDRLVSYYYFLRYGDNFRPHLVRKKHGDKMTFDECVEKSQPDCDPNNMWLQVPFFCGHAAECWKPGNHWALEQAKHNLVNHYLLVGVTEEMQDFITVLEATLPRFFKGATEYYLSSNKSHLRQTSSKVDPSLKTVERIQQSTVWKMENELYEFALEHFKFVKRKLLVKEANNVAQIYFYEKIRPK
Summary
Subunit
Homotrimer.
Similarity
Belongs to the sulfotransferase 3 family.
Keywords
Alternative splicing
Complete proteome
Disulfide bond
Glycoprotein
Golgi apparatus
Membrane
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain Heparin sulfate O-sulfotransferase
splice variant In isoform S4.
splice variant In isoform S4.
Uniprot
A0A2A4JFF5
A0A2H1W6L3
A0A194PZQ9
A0A194RBF4
H9J937
A0A212FCC2
+ More
A0A2W1BL67 A0A0K8TP20 A0A1Y1NAR3 A0A1B0Y0B6 A0A1S4F4P8 Q17FQ7 B4M966 A0A2J7QVT7 A0A067RNP2 E2C5N2 D6WJV8 A0A1Q3FB70 A0A1L8E4P5 A0A1L8E593 W8BM09 A0A087U5T2 A0A182YSP0 B4KK65 E2AC79 A0A182MW43 A0A0C9RGG6 A0A182W6Y7 A0A034W043 A0A182SPW5 A0A1B6C0H2 A0A0A1WSK7 A0A0K8V7F9 E0VPR1 A0A182JKZ9 A0A182RCQ8 B0WGK2 A0A1A9V7H3 A0A182GWJ3 A0A182QL53 A0A182JQG3 A0A026WUY6 Q29K51 A0A0M4E6I6 A0A084W834 A0A195ESL4 A0A195AT85 A0A1A9Z5N9 F4W7L0 A0A182WTY2 A0A182VJF8 A0A182HUJ8 A0A336MAC3 A0A158NPN7 A0A182UJM4 B4MYQ3 A0A0L0BU72 B4JP82 A0A182PPM9 A0A151WJ66 A0A232FCG7 Q7Q428 A0A2M4AUV4 A0A2M4AUP2 A0A182F2J1 K7ITS5 B3NM55 A0A1W4WYF9 A0A1B0BH17 A0A154PDL8 W5JK51 B3MK01 J3JYF7 A0A0N0BCJ5 A0A0L7R7E2 B4Q9Y1 A0A3B0JYH0 A0A1I8QBE3 B4PAY6 A0A088A780 P25722 P25722-2 B4I5V2 A0A1B6JKF4 A0A0A9VS21 A0A146LU86 A0A1S4EGM9 A0A3P9PHP7 A0A131XDT2 A0A195EM22 A0A1S3KC26 E9IY37 E9FUJ1 A0A3B5R5N7 A0A336N5V7 A0A3B3Y396 A0A3B3WFC0 A0A087X334 A0A3B3TXT1 A0A1A8QJ88
A0A2W1BL67 A0A0K8TP20 A0A1Y1NAR3 A0A1B0Y0B6 A0A1S4F4P8 Q17FQ7 B4M966 A0A2J7QVT7 A0A067RNP2 E2C5N2 D6WJV8 A0A1Q3FB70 A0A1L8E4P5 A0A1L8E593 W8BM09 A0A087U5T2 A0A182YSP0 B4KK65 E2AC79 A0A182MW43 A0A0C9RGG6 A0A182W6Y7 A0A034W043 A0A182SPW5 A0A1B6C0H2 A0A0A1WSK7 A0A0K8V7F9 E0VPR1 A0A182JKZ9 A0A182RCQ8 B0WGK2 A0A1A9V7H3 A0A182GWJ3 A0A182QL53 A0A182JQG3 A0A026WUY6 Q29K51 A0A0M4E6I6 A0A084W834 A0A195ESL4 A0A195AT85 A0A1A9Z5N9 F4W7L0 A0A182WTY2 A0A182VJF8 A0A182HUJ8 A0A336MAC3 A0A158NPN7 A0A182UJM4 B4MYQ3 A0A0L0BU72 B4JP82 A0A182PPM9 A0A151WJ66 A0A232FCG7 Q7Q428 A0A2M4AUV4 A0A2M4AUP2 A0A182F2J1 K7ITS5 B3NM55 A0A1W4WYF9 A0A1B0BH17 A0A154PDL8 W5JK51 B3MK01 J3JYF7 A0A0N0BCJ5 A0A0L7R7E2 B4Q9Y1 A0A3B0JYH0 A0A1I8QBE3 B4PAY6 A0A088A780 P25722 P25722-2 B4I5V2 A0A1B6JKF4 A0A0A9VS21 A0A146LU86 A0A1S4EGM9 A0A3P9PHP7 A0A131XDT2 A0A195EM22 A0A1S3KC26 E9IY37 E9FUJ1 A0A3B5R5N7 A0A336N5V7 A0A3B3Y396 A0A3B3WFC0 A0A087X334 A0A3B3TXT1 A0A1A8QJ88
EC Number
2.8.2.-
Pubmed
26354079
19121390
22118469
28756777
26369729
28004739
+ More
17510324 17994087 24845553 20798317 18362917 19820115 24495485 25244985 25348373 25830018 20566863 26483478 24508170 30249741 15632085 24438588 21719571 21347285 26108605 28648823 12364791 14747013 17210077 20075255 20920257 23761445 22516182 22936249 17550304 1936954 10731132 12537572 12537569 25401762 26823975 28049606 21282665 21292972 23542700
17510324 17994087 24845553 20798317 18362917 19820115 24495485 25244985 25348373 25830018 20566863 26483478 24508170 30249741 15632085 24438588 21719571 21347285 26108605 28648823 12364791 14747013 17210077 20075255 20920257 23761445 22516182 22936249 17550304 1936954 10731132 12537572 12537569 25401762 26823975 28049606 21282665 21292972 23542700
EMBL
NWSH01001783
PCG70153.1
ODYU01006354
SOQ48134.1
KQ459584
KPI98523.1
+ More
KQ460644 KPJ13231.1 BABH01004468 AGBW02009207 OWR51381.1 KZ150052 PZC74384.1 GDAI01001690 JAI15913.1 GEZM01010621 JAV93970.1 KT000407 ANI86005.1 CH477269 EAT45367.1 CH940654 EDW57742.2 NEVH01009771 PNF32698.1 KK852514 KDR22215.1 GL452776 EFN76743.1 KQ971342 EFA04624.1 GFDL01010283 JAV24762.1 GFDF01000383 JAV13701.1 GFDF01000382 JAV13702.1 GAMC01006723 JAB99832.1 KK118321 KFM72721.1 CH933807 EDW11583.2 GL438428 EFN68973.1 AXCM01004769 GBYB01012205 GBYB01012207 JAG81972.1 JAG81974.1 GAKP01011447 JAC47505.1 GEDC01030300 JAS06998.1 GBXI01012889 JAD01403.1 GDHF01017626 JAI34688.1 DS235371 EEB15367.1 DS231927 EDS27031.1 JXUM01093395 JXUM01093396 KQ564054 KXJ72861.1 AXCN02000760 KK107105 QOIP01000011 EZA59476.1 RLU16804.1 CH379061 EAL33327.2 CP012523 ALC39971.1 ATLV01021355 KE525317 KFB46378.1 KQ981986 KYN31218.1 KQ976745 KYM75406.1 GL887844 EGI69883.1 APCN01002465 UFQT01000300 SSX22988.1 ADTU01022666 CH963913 EDW77242.1 JRES01001455 KNC22764.1 CH916372 EDV98712.1 KQ983049 KYQ47871.1 NNAY01000473 OXU28138.1 AAAB01008964 EAA12392.4 GGFK01011171 MBW44492.1 GGFK01011170 MBW44491.1 CH954179 EDV54655.1 JXJN01014109 JXJN01014110 KQ434869 KZC09348.1 ADMH02000888 ETN64752.1 CH902620 EDV32456.1 BT128283 AEE63243.1 KQ435916 KOX68798.1 KQ414642 KOC66753.1 CM000361 CM002910 EDX05511.1 KMY91014.1 OUUW01000010 SPP85482.1 CM000158 EDW90415.1 X60218 AE014134 AY058422 CH480822 EDW55758.1 GECU01008036 JAS99670.1 GBHO01045528 JAF98075.1 GDHC01008679 JAQ09950.1 GEFH01004256 JAP64325.1 KQ978730 KYN28972.1 GL766903 EFZ14513.1 GL732525 EFX88915.1 UFQT01002680 SSX33928.1 AYCK01017732 HAEG01012904 SBR93546.1
KQ460644 KPJ13231.1 BABH01004468 AGBW02009207 OWR51381.1 KZ150052 PZC74384.1 GDAI01001690 JAI15913.1 GEZM01010621 JAV93970.1 KT000407 ANI86005.1 CH477269 EAT45367.1 CH940654 EDW57742.2 NEVH01009771 PNF32698.1 KK852514 KDR22215.1 GL452776 EFN76743.1 KQ971342 EFA04624.1 GFDL01010283 JAV24762.1 GFDF01000383 JAV13701.1 GFDF01000382 JAV13702.1 GAMC01006723 JAB99832.1 KK118321 KFM72721.1 CH933807 EDW11583.2 GL438428 EFN68973.1 AXCM01004769 GBYB01012205 GBYB01012207 JAG81972.1 JAG81974.1 GAKP01011447 JAC47505.1 GEDC01030300 JAS06998.1 GBXI01012889 JAD01403.1 GDHF01017626 JAI34688.1 DS235371 EEB15367.1 DS231927 EDS27031.1 JXUM01093395 JXUM01093396 KQ564054 KXJ72861.1 AXCN02000760 KK107105 QOIP01000011 EZA59476.1 RLU16804.1 CH379061 EAL33327.2 CP012523 ALC39971.1 ATLV01021355 KE525317 KFB46378.1 KQ981986 KYN31218.1 KQ976745 KYM75406.1 GL887844 EGI69883.1 APCN01002465 UFQT01000300 SSX22988.1 ADTU01022666 CH963913 EDW77242.1 JRES01001455 KNC22764.1 CH916372 EDV98712.1 KQ983049 KYQ47871.1 NNAY01000473 OXU28138.1 AAAB01008964 EAA12392.4 GGFK01011171 MBW44492.1 GGFK01011170 MBW44491.1 CH954179 EDV54655.1 JXJN01014109 JXJN01014110 KQ434869 KZC09348.1 ADMH02000888 ETN64752.1 CH902620 EDV32456.1 BT128283 AEE63243.1 KQ435916 KOX68798.1 KQ414642 KOC66753.1 CM000361 CM002910 EDX05511.1 KMY91014.1 OUUW01000010 SPP85482.1 CM000158 EDW90415.1 X60218 AE014134 AY058422 CH480822 EDW55758.1 GECU01008036 JAS99670.1 GBHO01045528 JAF98075.1 GDHC01008679 JAQ09950.1 GEFH01004256 JAP64325.1 KQ978730 KYN28972.1 GL766903 EFZ14513.1 GL732525 EFX88915.1 UFQT01002680 SSX33928.1 AYCK01017732 HAEG01012904 SBR93546.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000005204
UP000007151
UP000008820
+ More
UP000008792 UP000235965 UP000027135 UP000008237 UP000007266 UP000054359 UP000076408 UP000009192 UP000000311 UP000075883 UP000075920 UP000075901 UP000009046 UP000075880 UP000075900 UP000002320 UP000078200 UP000069940 UP000249989 UP000075886 UP000075881 UP000053097 UP000279307 UP000001819 UP000092553 UP000030765 UP000078541 UP000078540 UP000092445 UP000007755 UP000076407 UP000075903 UP000075840 UP000005205 UP000075902 UP000007798 UP000037069 UP000001070 UP000075885 UP000075809 UP000215335 UP000007062 UP000069272 UP000002358 UP000008711 UP000192223 UP000092460 UP000076502 UP000000673 UP000007801 UP000053105 UP000053825 UP000000304 UP000268350 UP000095300 UP000002282 UP000005203 UP000000803 UP000001292 UP000079169 UP000242638 UP000078492 UP000085678 UP000000305 UP000002852 UP000261480 UP000028760 UP000261500
UP000008792 UP000235965 UP000027135 UP000008237 UP000007266 UP000054359 UP000076408 UP000009192 UP000000311 UP000075883 UP000075920 UP000075901 UP000009046 UP000075880 UP000075900 UP000002320 UP000078200 UP000069940 UP000249989 UP000075886 UP000075881 UP000053097 UP000279307 UP000001819 UP000092553 UP000030765 UP000078541 UP000078540 UP000092445 UP000007755 UP000076407 UP000075903 UP000075840 UP000005205 UP000075902 UP000007798 UP000037069 UP000001070 UP000075885 UP000075809 UP000215335 UP000007062 UP000069272 UP000002358 UP000008711 UP000192223 UP000092460 UP000076502 UP000000673 UP000007801 UP000053105 UP000053825 UP000000304 UP000268350 UP000095300 UP000002282 UP000005203 UP000000803 UP000001292 UP000079169 UP000242638 UP000078492 UP000085678 UP000000305 UP000002852 UP000261480 UP000028760 UP000261500
Interpro
ProteinModelPortal
A0A2A4JFF5
A0A2H1W6L3
A0A194PZQ9
A0A194RBF4
H9J937
A0A212FCC2
+ More
A0A2W1BL67 A0A0K8TP20 A0A1Y1NAR3 A0A1B0Y0B6 A0A1S4F4P8 Q17FQ7 B4M966 A0A2J7QVT7 A0A067RNP2 E2C5N2 D6WJV8 A0A1Q3FB70 A0A1L8E4P5 A0A1L8E593 W8BM09 A0A087U5T2 A0A182YSP0 B4KK65 E2AC79 A0A182MW43 A0A0C9RGG6 A0A182W6Y7 A0A034W043 A0A182SPW5 A0A1B6C0H2 A0A0A1WSK7 A0A0K8V7F9 E0VPR1 A0A182JKZ9 A0A182RCQ8 B0WGK2 A0A1A9V7H3 A0A182GWJ3 A0A182QL53 A0A182JQG3 A0A026WUY6 Q29K51 A0A0M4E6I6 A0A084W834 A0A195ESL4 A0A195AT85 A0A1A9Z5N9 F4W7L0 A0A182WTY2 A0A182VJF8 A0A182HUJ8 A0A336MAC3 A0A158NPN7 A0A182UJM4 B4MYQ3 A0A0L0BU72 B4JP82 A0A182PPM9 A0A151WJ66 A0A232FCG7 Q7Q428 A0A2M4AUV4 A0A2M4AUP2 A0A182F2J1 K7ITS5 B3NM55 A0A1W4WYF9 A0A1B0BH17 A0A154PDL8 W5JK51 B3MK01 J3JYF7 A0A0N0BCJ5 A0A0L7R7E2 B4Q9Y1 A0A3B0JYH0 A0A1I8QBE3 B4PAY6 A0A088A780 P25722 P25722-2 B4I5V2 A0A1B6JKF4 A0A0A9VS21 A0A146LU86 A0A1S4EGM9 A0A3P9PHP7 A0A131XDT2 A0A195EM22 A0A1S3KC26 E9IY37 E9FUJ1 A0A3B5R5N7 A0A336N5V7 A0A3B3Y396 A0A3B3WFC0 A0A087X334 A0A3B3TXT1 A0A1A8QJ88
A0A2W1BL67 A0A0K8TP20 A0A1Y1NAR3 A0A1B0Y0B6 A0A1S4F4P8 Q17FQ7 B4M966 A0A2J7QVT7 A0A067RNP2 E2C5N2 D6WJV8 A0A1Q3FB70 A0A1L8E4P5 A0A1L8E593 W8BM09 A0A087U5T2 A0A182YSP0 B4KK65 E2AC79 A0A182MW43 A0A0C9RGG6 A0A182W6Y7 A0A034W043 A0A182SPW5 A0A1B6C0H2 A0A0A1WSK7 A0A0K8V7F9 E0VPR1 A0A182JKZ9 A0A182RCQ8 B0WGK2 A0A1A9V7H3 A0A182GWJ3 A0A182QL53 A0A182JQG3 A0A026WUY6 Q29K51 A0A0M4E6I6 A0A084W834 A0A195ESL4 A0A195AT85 A0A1A9Z5N9 F4W7L0 A0A182WTY2 A0A182VJF8 A0A182HUJ8 A0A336MAC3 A0A158NPN7 A0A182UJM4 B4MYQ3 A0A0L0BU72 B4JP82 A0A182PPM9 A0A151WJ66 A0A232FCG7 Q7Q428 A0A2M4AUV4 A0A2M4AUP2 A0A182F2J1 K7ITS5 B3NM55 A0A1W4WYF9 A0A1B0BH17 A0A154PDL8 W5JK51 B3MK01 J3JYF7 A0A0N0BCJ5 A0A0L7R7E2 B4Q9Y1 A0A3B0JYH0 A0A1I8QBE3 B4PAY6 A0A088A780 P25722 P25722-2 B4I5V2 A0A1B6JKF4 A0A0A9VS21 A0A146LU86 A0A1S4EGM9 A0A3P9PHP7 A0A131XDT2 A0A195EM22 A0A1S3KC26 E9IY37 E9FUJ1 A0A3B5R5N7 A0A336N5V7 A0A3B3Y396 A0A3B3WFC0 A0A087X334 A0A3B3TXT1 A0A1A8QJ88
PDB
3F5F
E-value=5.25974e-106,
Score=981
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
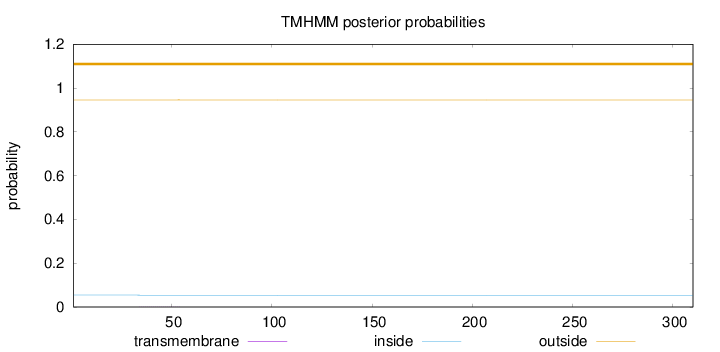
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00807999999999996
Exp number, first 60 AAs:
0.00567
Total prob of N-in:
0.05432
outside
1 - 310
Population Genetic Test Statistics
Pi
193.06916
Theta
204.07914
Tajima's D
-0.165932
CLR
172.583034
CSRT
0.330083495825209
Interpretation
Uncertain
