Gene
KWMTBOMO01694 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006032
Annotation
PREDICTED:_adenylate_kinase_isoenzyme_1_isoform_X3_[Bombyx_mori]
Full name
Adenylate kinase isoenzyme 1
Alternative Name
ATP-AMP transphosphorylase 1
ATP:AMP phosphotransferase
Adenylate monophosphate kinase
Myokinase
ATP:AMP phosphotransferase
Adenylate monophosphate kinase
Myokinase
Location in the cell
Nuclear Reliability : 4.258
Sequence
CDS
ATGGCACCCATAGATACATCGAAAAAGTCAATCGTGTGGGTCCTGGGAGGTCCCGGATCAGGAAAGGGAACACAATGCGACAAGATCGTCGCCAAATACGGGTTCACTCACCTCTCGACTGGAGACCTGCTCAGGGCTGAAGTCAAAAGCGGCTCGGAGAGGGCGAAATGCCTTACCGCTATTATGGAAAGAGGCGAGCTTGTCCCTAATGAAATCGTCCTGGATCTGCTAAAAGAAGCGATCCTGGCCCGCGCTGAAGAATCCAAAGGCTTCCTCATCGACGGCTACCCTCGCGAGAAGTCACAGGGTATCGCTTTTGAAGAGAACATCGCTCCTGTTTCGGTAATAATATACTTCGAGGCCTCCTCGGAGACGCTGACGCAGCGCCTGCTGGGCCGCGCCGCCAGCTCCGGCCGTGCCGATGACAACGAGGAGACCATCAAGCTCAGGCTGAAGACCTTCCTGGACAACAACGACCTCGTACTGGCTCAATACCCGGACAAACTCAGGAGGATAAACGCGGAACGTCCTGTGGATGAGATCTTCGCGGAGGTACAGTCGATCCTGGACCCGCTGGTCGCGTGA
Protein
MAPIDTSKKSIVWVLGGPGSGKGTQCDKIVAKYGFTHLSTGDLLRAEVKSGSERAKCLTAIMERGELVPNEIVLDLLKEAILARAEESKGFLIDGYPREKSQGIAFEENIAPVSVIIYFEASSETLTQRLLGRAASSGRADDNEETIKLRLKTFLDNNDLVLAQYPDKLRRINAERPVDEIFAEVQSILDPLVA
Summary
Description
Catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP. Also displays broad nucleoside diphosphate kinase activity. Plays an important role in cellular energy homeostasis and in adenine nucleotide metabolism.
Catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP. Also possesses broad nucleoside diphosphate kinase activity. Plays an important role in cellular energy homeostasis and in adenine nucleotide metabolism (By similarity). May provide a mechanism to buffer the adenylate energy charge for sperm motility.
Catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP. Also possesses broad nucleoside diphosphate kinase activity. Plays an important role in cellular energy homeostasis and in adenine nucleotide metabolism (By similarity). May provide a mechanism to buffer the adenylate energy charge for sperm motility.
Catalytic Activity
AMP + ATP = 2 ADP
a 2'-deoxyribonucleoside 5'-diphosphate + ATP = a 2'-deoxyribonucleoside 5'-triphosphate + ADP
a ribonucleoside 5'-diphosphate + ATP = a ribonucleoside 5'-triphosphate + ADP
a 2'-deoxyribonucleoside 5'-diphosphate + ATP = a 2'-deoxyribonucleoside 5'-triphosphate + ADP
a ribonucleoside 5'-diphosphate + ATP = a ribonucleoside 5'-triphosphate + ADP
Subunit
Monomer.
Similarity
Belongs to the adenylate kinase family.
Belongs to the adenylate kinase family. AK1 subfamily.
Belongs to the adenylate kinase family. AK1 subfamily.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Kinase
Nucleotide-binding
Reference proteome
Transferase
Acetylation
Alternative splicing
Direct protein sequencing
Phosphoprotein
Feature
chain Adenylate kinase isoenzyme 1
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J940
A0A220K8L6
A0A194R5Y6
A0A194PYW9
A0A2W1BNE1
A0A2A4KB60
+ More
A0A2A4KAT5 A0A2H1VQ92 D6WJX7 J3JVF3 U4U9W1 A0A0T6B682 A0A1A9W548 A0A1Y1N9H6 A0A0P4WDJ8 A0A034WMG6 A0A034WQY9 A0A0K8UAD9 A0A0K8V9S7 W8CAY2 C3Y327 W8BWF8 A0A0A1WER3 A0A0A1WKU5 C1BPN7 C3YAN3 A0A212EJ57 A0A0D3L709 H3BB97 H3BB96 A0A2H8TET5 Q6GQF9 Q6DFN0 F7CEM0 C1BTY0 K4FYQ8 P05081 T1FNK1 A0A1B6MUT7 A0A2S2P5H6 A0A2W1BM84 A0A1B6LHM9 R4WDG3 A0A336MQY4 A0A310TQK3 Q66J33 A0A2D4Q353 A0A067REE0 A0A1B6M5G1 J9JQL8 Q9R0Y5 A0A182K5K4 V4ANU9 A0A0V0G4J5 C1BZW4 A0A2D4JGP1 A0A2H6NA96 A0A226NXC1 A0A023EKD2 E2B248 E9H0V7 A0A1U7RV51 A0A023EKE5 A0A1B6HEU2 Q9R0Y5-2 A0A2P4SF41 T1HFU1 Q16W51 F6PLC0 Q16W53 A0A1U7S5Y0 W5M844 F4WX98 A0A182J6U4 A0A1U7RHU6 A0A1S4H0P8 Q7QGG2 A0A084VM63 A0A0P5EI24 A0A224XKK9 G1QBS7 A0A0A9Y5Y3 A0A0K8SFI8 A0A069DX59 A0A151WNH3 P39069 A0A3Q0CFS5 A0A287D6D9 A0A3M7RII3 A0A0P4VJV2 A0A1S4H0S9 S7Q8G6 A0A1A6GV92 A0A1I8NPD1 T1E2E3 I3N0J3 K7FSU9 A0A1I8NPD9 G1PN10 A0A026WP83
A0A2A4KAT5 A0A2H1VQ92 D6WJX7 J3JVF3 U4U9W1 A0A0T6B682 A0A1A9W548 A0A1Y1N9H6 A0A0P4WDJ8 A0A034WMG6 A0A034WQY9 A0A0K8UAD9 A0A0K8V9S7 W8CAY2 C3Y327 W8BWF8 A0A0A1WER3 A0A0A1WKU5 C1BPN7 C3YAN3 A0A212EJ57 A0A0D3L709 H3BB97 H3BB96 A0A2H8TET5 Q6GQF9 Q6DFN0 F7CEM0 C1BTY0 K4FYQ8 P05081 T1FNK1 A0A1B6MUT7 A0A2S2P5H6 A0A2W1BM84 A0A1B6LHM9 R4WDG3 A0A336MQY4 A0A310TQK3 Q66J33 A0A2D4Q353 A0A067REE0 A0A1B6M5G1 J9JQL8 Q9R0Y5 A0A182K5K4 V4ANU9 A0A0V0G4J5 C1BZW4 A0A2D4JGP1 A0A2H6NA96 A0A226NXC1 A0A023EKD2 E2B248 E9H0V7 A0A1U7RV51 A0A023EKE5 A0A1B6HEU2 Q9R0Y5-2 A0A2P4SF41 T1HFU1 Q16W51 F6PLC0 Q16W53 A0A1U7S5Y0 W5M844 F4WX98 A0A182J6U4 A0A1U7RHU6 A0A1S4H0P8 Q7QGG2 A0A084VM63 A0A0P5EI24 A0A224XKK9 G1QBS7 A0A0A9Y5Y3 A0A0K8SFI8 A0A069DX59 A0A151WNH3 P39069 A0A3Q0CFS5 A0A287D6D9 A0A3M7RII3 A0A0P4VJV2 A0A1S4H0S9 S7Q8G6 A0A1A6GV92 A0A1I8NPD1 T1E2E3 I3N0J3 K7FSU9 A0A1I8NPD9 G1PN10 A0A026WP83
EC Number
2.7.4.3
2.7.4.6
2.7.4.6
Pubmed
19121390
28630352
26354079
28756777
18362917
19820115
+ More
22516182 23537049 28004739 25348373 24495485 18563158 25830018 22118469 9215903 12454917 12477932 17495919 23056606 2419325 2844738 2161682 23254933 23691247 24845553 10557075 16790685 16141072 19468303 15489334 16452087 21183079 24621616 24945155 20798317 21292972 17510324 20431018 21719571 12364791 24438588 21993624 25401762 26334808 8468325 22673903 30375419 27129103 24330624 17381049 24508170 30249741
22516182 23537049 28004739 25348373 24495485 18563158 25830018 22118469 9215903 12454917 12477932 17495919 23056606 2419325 2844738 2161682 23254933 23691247 24845553 10557075 16790685 16141072 19468303 15489334 16452087 21183079 24621616 24945155 20798317 21292972 17510324 20431018 21719571 12364791 24438588 21993624 25401762 26334808 8468325 22673903 30375419 27129103 24330624 17381049 24508170 30249741
EMBL
BABH01004466
MF319716
ASJ26430.1
KQ460644
KPJ13233.1
KQ459584
+ More
KPI98521.1 KZ150052 PZC74386.1 NWSH01000011 PCG80902.1 PCG80903.1 ODYU01003780 SOQ42967.1 KQ971342 EFA03917.1 APGK01043872 BT127221 KB741018 AEE62183.1 ENN75232.1 KB632152 ERL89138.1 LJIG01009533 KRT82889.1 GEZM01009463 JAV94572.1 GDRN01071336 GDRN01071334 JAI63707.1 GAKP01002236 JAC56716.1 GAKP01002235 JAC56717.1 GDHF01028672 JAI23642.1 GDHF01016605 JAI35709.1 GAMC01005306 JAC01250.1 GG666482 EEN65439.1 GAMC01005307 JAC01249.1 GBXI01016793 JAC97498.1 GBXI01014633 JAC99658.1 BT076566 ACO10990.1 GG666494 EEN62781.1 AGBW02014536 OWR41498.1 KC822758 AHJ80786.1 AFYH01046138 GFXV01000812 MBW12617.1 BC072785 AAH72785.1 BC076704 CR855821 AAH76704.1 CAJ83224.1 BT078059 BT120860 BT121139 HACA01028991 ACO12483.1 ADD24500.1 ADD38069.1 CDW46352.1 JX053354 AFK11582.1 M12153 D00251 AMQM01007061 KB097558 ESN94879.1 GEBQ01000273 JAT39704.1 GGMR01011983 MBY24602.1 PZC74387.1 GEBQ01026828 GEBQ01016755 GEBQ01009572 JAT13149.1 JAT23222.1 JAT30405.1 AK417663 BAN20878.1 UFQS01000997 UFQT01000997 SSX08345.1 SSX28368.1 KV467239 OCT57008.1 BC081078 AAH81078.1 IACN01117612 LAB63899.1 KK852514 KDR22221.1 GEBQ01008825 JAT31152.1 ABLF02039069 AJ010108 AJ010109 DQ486026 AK046613 AK089270 AL772271 BC014802 BC054366 KB201205 ESO98837.1 GECL01003520 JAP02604.1 BT080143 ACO14567.1 IACK01186307 LAA95676.1 IACI01077941 LAA28108.1 AWGT02000307 OXB72144.1 GAPW01003990 JAC09608.1 GL445059 EFN60223.1 GL732582 EFX74525.1 GAPW01003991 JAC09607.1 GECU01034520 GECU01031332 GECU01025307 GECU01022044 GECU01020516 GECU01014224 GECU01012683 JAS73186.1 JAS76374.1 JAS82399.1 JAS85662.1 JAS87190.1 JAS93482.1 JAS95023.1 PPHD01055425 POI22721.1 ACPB03001435 CH477575 EAT38802.2 AAMC01047190 AAMC01047191 AAMC01047192 AAMC01047193 AAMC01047194 AAMC01047195 EAT38800.1 EAT38801.1 AHAT01025332 GL888423 EGI61147.1 AAAB01008835 EAA05792.3 ATLV01014580 KE524975 KFB39057.1 GDIP01146162 JAJ77240.1 GFTR01003419 JAW13007.1 AAPE02044248 GBHO01016030 GBHO01016029 JAG27574.1 JAG27575.1 GBRD01013779 GBRD01013778 GBRD01013777 JAG52047.1 GBGD01002970 JAC85919.1 KQ982926 KYQ49235.1 AB022701 BC089797 D13376 AGTP01020068 REGN01003329 RNA23200.1 GDKW01002078 JAI54517.1 KE164341 EPQ17187.1 LZPO01073204 OBS69257.1 GALA01000799 JAA94053.1 AGCU01057997 AGCU01057998 AGCU01057999 AGCU01058000 AGCU01058001 AGCU01058002 AGCU01058003 AGCU01058004 AGCU01058005 AGCU01058006 KK107154 QOIP01000001 EZA56909.1 RLU27201.1
KPI98521.1 KZ150052 PZC74386.1 NWSH01000011 PCG80902.1 PCG80903.1 ODYU01003780 SOQ42967.1 KQ971342 EFA03917.1 APGK01043872 BT127221 KB741018 AEE62183.1 ENN75232.1 KB632152 ERL89138.1 LJIG01009533 KRT82889.1 GEZM01009463 JAV94572.1 GDRN01071336 GDRN01071334 JAI63707.1 GAKP01002236 JAC56716.1 GAKP01002235 JAC56717.1 GDHF01028672 JAI23642.1 GDHF01016605 JAI35709.1 GAMC01005306 JAC01250.1 GG666482 EEN65439.1 GAMC01005307 JAC01249.1 GBXI01016793 JAC97498.1 GBXI01014633 JAC99658.1 BT076566 ACO10990.1 GG666494 EEN62781.1 AGBW02014536 OWR41498.1 KC822758 AHJ80786.1 AFYH01046138 GFXV01000812 MBW12617.1 BC072785 AAH72785.1 BC076704 CR855821 AAH76704.1 CAJ83224.1 BT078059 BT120860 BT121139 HACA01028991 ACO12483.1 ADD24500.1 ADD38069.1 CDW46352.1 JX053354 AFK11582.1 M12153 D00251 AMQM01007061 KB097558 ESN94879.1 GEBQ01000273 JAT39704.1 GGMR01011983 MBY24602.1 PZC74387.1 GEBQ01026828 GEBQ01016755 GEBQ01009572 JAT13149.1 JAT23222.1 JAT30405.1 AK417663 BAN20878.1 UFQS01000997 UFQT01000997 SSX08345.1 SSX28368.1 KV467239 OCT57008.1 BC081078 AAH81078.1 IACN01117612 LAB63899.1 KK852514 KDR22221.1 GEBQ01008825 JAT31152.1 ABLF02039069 AJ010108 AJ010109 DQ486026 AK046613 AK089270 AL772271 BC014802 BC054366 KB201205 ESO98837.1 GECL01003520 JAP02604.1 BT080143 ACO14567.1 IACK01186307 LAA95676.1 IACI01077941 LAA28108.1 AWGT02000307 OXB72144.1 GAPW01003990 JAC09608.1 GL445059 EFN60223.1 GL732582 EFX74525.1 GAPW01003991 JAC09607.1 GECU01034520 GECU01031332 GECU01025307 GECU01022044 GECU01020516 GECU01014224 GECU01012683 JAS73186.1 JAS76374.1 JAS82399.1 JAS85662.1 JAS87190.1 JAS93482.1 JAS95023.1 PPHD01055425 POI22721.1 ACPB03001435 CH477575 EAT38802.2 AAMC01047190 AAMC01047191 AAMC01047192 AAMC01047193 AAMC01047194 AAMC01047195 EAT38800.1 EAT38801.1 AHAT01025332 GL888423 EGI61147.1 AAAB01008835 EAA05792.3 ATLV01014580 KE524975 KFB39057.1 GDIP01146162 JAJ77240.1 GFTR01003419 JAW13007.1 AAPE02044248 GBHO01016030 GBHO01016029 JAG27574.1 JAG27575.1 GBRD01013779 GBRD01013778 GBRD01013777 JAG52047.1 GBGD01002970 JAC85919.1 KQ982926 KYQ49235.1 AB022701 BC089797 D13376 AGTP01020068 REGN01003329 RNA23200.1 GDKW01002078 JAI54517.1 KE164341 EPQ17187.1 LZPO01073204 OBS69257.1 GALA01000799 JAA94053.1 AGCU01057997 AGCU01057998 AGCU01057999 AGCU01058000 AGCU01058001 AGCU01058002 AGCU01058003 AGCU01058004 AGCU01058005 AGCU01058006 KK107154 QOIP01000001 EZA56909.1 RLU27201.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007266
UP000019118
+ More
UP000030742 UP000091820 UP000001554 UP000007151 UP000008672 UP000002280 UP000000539 UP000015101 UP000027135 UP000007819 UP000000589 UP000075881 UP000030746 UP000198419 UP000000311 UP000000305 UP000189705 UP000015103 UP000008820 UP000008143 UP000018468 UP000007755 UP000075880 UP000189706 UP000007062 UP000030765 UP000001074 UP000075809 UP000002494 UP000005215 UP000276133 UP000092124 UP000095300 UP000007267 UP000053097 UP000279307
UP000030742 UP000091820 UP000001554 UP000007151 UP000008672 UP000002280 UP000000539 UP000015101 UP000027135 UP000007819 UP000000589 UP000075881 UP000030746 UP000198419 UP000000311 UP000000305 UP000189705 UP000015103 UP000008820 UP000008143 UP000018468 UP000007755 UP000075880 UP000189706 UP000007062 UP000030765 UP000001074 UP000075809 UP000002494 UP000005215 UP000276133 UP000092124 UP000095300 UP000007267 UP000053097 UP000279307
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9J940
A0A220K8L6
A0A194R5Y6
A0A194PYW9
A0A2W1BNE1
A0A2A4KB60
+ More
A0A2A4KAT5 A0A2H1VQ92 D6WJX7 J3JVF3 U4U9W1 A0A0T6B682 A0A1A9W548 A0A1Y1N9H6 A0A0P4WDJ8 A0A034WMG6 A0A034WQY9 A0A0K8UAD9 A0A0K8V9S7 W8CAY2 C3Y327 W8BWF8 A0A0A1WER3 A0A0A1WKU5 C1BPN7 C3YAN3 A0A212EJ57 A0A0D3L709 H3BB97 H3BB96 A0A2H8TET5 Q6GQF9 Q6DFN0 F7CEM0 C1BTY0 K4FYQ8 P05081 T1FNK1 A0A1B6MUT7 A0A2S2P5H6 A0A2W1BM84 A0A1B6LHM9 R4WDG3 A0A336MQY4 A0A310TQK3 Q66J33 A0A2D4Q353 A0A067REE0 A0A1B6M5G1 J9JQL8 Q9R0Y5 A0A182K5K4 V4ANU9 A0A0V0G4J5 C1BZW4 A0A2D4JGP1 A0A2H6NA96 A0A226NXC1 A0A023EKD2 E2B248 E9H0V7 A0A1U7RV51 A0A023EKE5 A0A1B6HEU2 Q9R0Y5-2 A0A2P4SF41 T1HFU1 Q16W51 F6PLC0 Q16W53 A0A1U7S5Y0 W5M844 F4WX98 A0A182J6U4 A0A1U7RHU6 A0A1S4H0P8 Q7QGG2 A0A084VM63 A0A0P5EI24 A0A224XKK9 G1QBS7 A0A0A9Y5Y3 A0A0K8SFI8 A0A069DX59 A0A151WNH3 P39069 A0A3Q0CFS5 A0A287D6D9 A0A3M7RII3 A0A0P4VJV2 A0A1S4H0S9 S7Q8G6 A0A1A6GV92 A0A1I8NPD1 T1E2E3 I3N0J3 K7FSU9 A0A1I8NPD9 G1PN10 A0A026WP83
A0A2A4KAT5 A0A2H1VQ92 D6WJX7 J3JVF3 U4U9W1 A0A0T6B682 A0A1A9W548 A0A1Y1N9H6 A0A0P4WDJ8 A0A034WMG6 A0A034WQY9 A0A0K8UAD9 A0A0K8V9S7 W8CAY2 C3Y327 W8BWF8 A0A0A1WER3 A0A0A1WKU5 C1BPN7 C3YAN3 A0A212EJ57 A0A0D3L709 H3BB97 H3BB96 A0A2H8TET5 Q6GQF9 Q6DFN0 F7CEM0 C1BTY0 K4FYQ8 P05081 T1FNK1 A0A1B6MUT7 A0A2S2P5H6 A0A2W1BM84 A0A1B6LHM9 R4WDG3 A0A336MQY4 A0A310TQK3 Q66J33 A0A2D4Q353 A0A067REE0 A0A1B6M5G1 J9JQL8 Q9R0Y5 A0A182K5K4 V4ANU9 A0A0V0G4J5 C1BZW4 A0A2D4JGP1 A0A2H6NA96 A0A226NXC1 A0A023EKD2 E2B248 E9H0V7 A0A1U7RV51 A0A023EKE5 A0A1B6HEU2 Q9R0Y5-2 A0A2P4SF41 T1HFU1 Q16W51 F6PLC0 Q16W53 A0A1U7S5Y0 W5M844 F4WX98 A0A182J6U4 A0A1U7RHU6 A0A1S4H0P8 Q7QGG2 A0A084VM63 A0A0P5EI24 A0A224XKK9 G1QBS7 A0A0A9Y5Y3 A0A0K8SFI8 A0A069DX59 A0A151WNH3 P39069 A0A3Q0CFS5 A0A287D6D9 A0A3M7RII3 A0A0P4VJV2 A0A1S4H0S9 S7Q8G6 A0A1A6GV92 A0A1I8NPD1 T1E2E3 I3N0J3 K7FSU9 A0A1I8NPD9 G1PN10 A0A026WP83
PDB
5XZ2
E-value=6.33619e-55,
Score=537
Ontologies
PATHWAY
GO
GO:0019205
GO:0006139
GO:0005524
GO:0004017
GO:0005737
GO:0046034
GO:0046033
GO:0004550
GO:0009142
GO:0006172
GO:0001520
GO:0005829
GO:0007050
GO:0005886
GO:0006165
GO:0043005
GO:0036126
GO:0048471
GO:0010828
GO:0030017
GO:0046103
GO:0009041
GO:0006207
GO:0004127
GO:0006221
GO:0033574
GO:0021772
GO:0030182
GO:0014042
GO:0032355
GO:0021549
GO:0007517
GO:0042493
GO:0014823
GO:0046083
GO:0016301
GO:0006355
GO:0006508
GO:0006811
GO:0016020
GO:0005515
GO:0003779
GO:0016021
PANTHER
Topology
Subcellular location
Cytoplasm
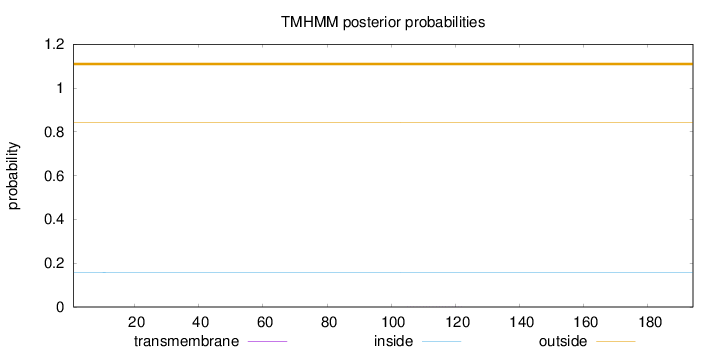
Length:
194
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00391
Exp number, first 60 AAs:
0.00052
Total prob of N-in:
0.15837
outside
1 - 194
Population Genetic Test Statistics
Pi
244.757614
Theta
179.35336
Tajima's D
1.657573
CLR
0.032589
CSRT
0.817159142042898
Interpretation
Uncertain
